













| General Information: |
|
| Name(s) found: |
PCK1 /
YKR097W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 549 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
gluconeogenesis
[IEP |
| Molecular Function: |
phosphoenolpyruvate carboxykinase (ATP) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPSKMNATV GSTSEVEQKI RQELALSDEV TTIRRNAPAA VLYEDGLKEN KTVISSSGAL 60
61 IAYSGVKTGR SPKDKRIVEE PTSKDEIWWG PVNKPCSERT WSINRERAAD YLRTRDHIYI 120
121 VDAFAGWDPK YRIKVRVVCA RAYHALFMTN MLIRPTEEEL AHFGEPDFTV WNAGQFPANL 180
181 HTQDMSSKST IEINFKAMEM IILGTEYAGE MKKGIFTVMF YLMPVHHNVL TLHSSANQGI 240
241 QNGDVTLFFG LSGTGKTTLS ADPHRLLIGD DEHCWSDHGV FNIEGGCYAK CINLSAEKEP 300
301 EIFDAIKFGS VLENVIYDEK SHVVDYDDSS ITENTRCAYP IDYIPSAKIP CLADSHPKNI 360
361 ILLTCDASGV LPPVSKLTPE QVMYHFISGY TSKMAGTEQG VTEPEPTFSS CFGQPFLALH 420
421 PIRYATMLAT KMSQHKANAY LINTGWTGSS YVSGGKRCPL KYTRAILDSI HDGSLANETY 480
481 ETLPIFNLQV PTKVNGVPAE LLNPAKNWSQ GESKYRGAVT NLANLFVQNF KIYQDRATPD 540
541 VLAAGPQFE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
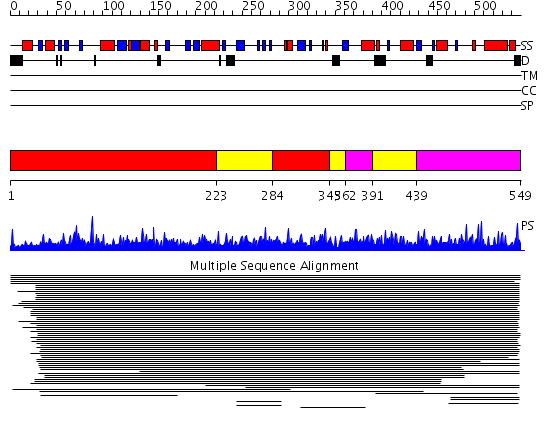
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] [284..344] |
2440.0 | Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-liase) | |
| 2 | View Details | [223..283] [345..361] [391..438] |
2440.0 | Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-liase) | |
| 3 | View Details | [362..390] [439..549] |
2440.0 | Phosphoenolpyruvate (PEP) carboxykinase (ATP-oxaloacetate carboxy-liase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)