













| General Information: |
|
| Name(s) found: |
BNA3 /
YJL060W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
NAD biosynthetic process
[IMP |
| Molecular Function: |
arylformamidase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQRFIRQFT NLMSTSRPKV VANKYFTSNT AKDVWSLTNE AAAKAANNSK NQGRELINLG 60
61 QGFFSYSPPQ FAIKEAQKAL DIPMVNQYSP TRGRPSLINS LIKLYSPIYN TELKAENVTV 120
121 TTGANEGILS CLMGLLNAGD EVIVFEPFFD QYIPNIELCG GKVVYVPINP PKELDQRNTR 180
181 GEEWTIDFEQ FEKAITSKTK AVIINTPHNP IGKVFTREEL TTLGNICVKH NVVIISDEVY 240
241 EHLYFTDSFT RIATLSPEIG QLTLTVGSAG KSFAATGWRI GWVLSLNAEL LSYAAKAHTR 300
301 ICFASPSPLQ EACANSINDA LKIGYFEKMR QEYINKFKIF TSIFDELGLP YTAPEGTYFV 360
361 LVDFSKVKIP EDYPYPEEIL NKGKDFRISH WLINELGVVA IPPTEFYIKE HEKAAENLLR 420
421 FAVCKDDAYL ENAVERLKLL KDYL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BNA3 CDC3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
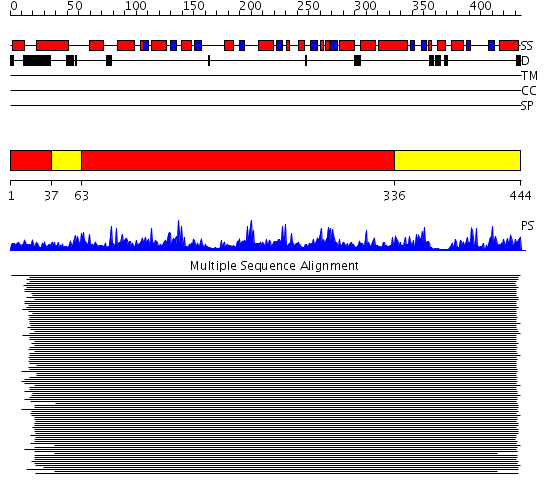
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..36] [63..335] |
482.9897 | Aromatic aminoacid aminotransferase, AroAT | |
| 2 | View Details | [37..62] [336..444] |
482.9897 | Aromatic aminoacid aminotransferase, AroAT |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)