













| General Information: |
|
| Name(s) found: |
SER33 /
YIL074C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 469 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
serine family amino acid biosynthetic process
[IMP |
| Molecular Function: |
phosphoglycerate dehydrogenase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYSAADNLQ DSFQRAMNFS GSPGAVSTSP TQSFMNTLPR RVSITKQPKA LKPFSTGDMN 60
61 ILLLENVNAT AIKIFKDQGY QVEFHKSSLP EDELIEKIKD VHAIGIRSKT RLTEKILQHA 120
121 RNLVCIGCFC IGTNQVDLKY AASKGIAVFN SPFSNSRSVA ELVIGEIISL ARQLGDRSIE 180
181 LHTGTWNKVA ARCWEVRGKT LGIIGYGHIG SQLSVLAEAM GLHVLYYDIV TIMALGTARQ 240
241 VSTLDELLNK SDFVTLHVPA TPETEKMLSA PQFAAMKDGA YVINASRGTV VDIPSLIQAV 300
301 KANKIAGAAL DVYPHEPAKN GEGSFNDELN SWTSELVSLP NIILTPHIGG STEEAQSSIG 360
361 IEVATALSKY INEGNSVGSV NFPEVSLKSL DYDQENTVRV LYIHRNVPGV LKTVNDILSD 420
421 HNIEKQFSDS HGEIAYLMAD ISSVNQSEIK DIYEKLNQTS AKVSIRLLY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SER3 SER33 |
| View Details | Krogan NJ, et al. (2006) |

|
SER3 SER33 |
| View Details | Gavin AC, et al. (2002) |
|
PHO12 SER3 SER33 |
| View Details | Qiu et al. (2008) |

|
SER3 SER33 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
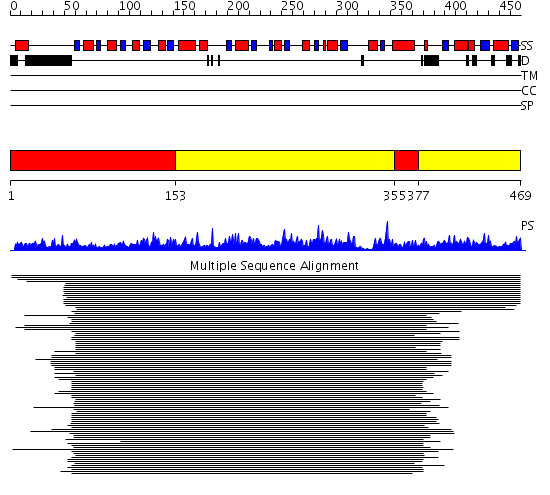
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] [355..376] |
1164.0 | Phosphoglycerate dehydrogenase; Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain | |
| 2 | View Details | [153..354] [377..469] |
1164.0 | Phosphoglycerate dehydrogenase; Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)