













| General Information: |
|
| Name(s) found: |
MIG2 /
YGL209W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 382 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter by glucose
[IGI |
| Molecular Function: |
sequence-specific DNA binding
[IDA specific transcriptional repressor activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKKQTNFPV DNENRPFRCD TCHRGFHRLE HKKRHLRTHT GEKPHHCAFP GCGKSFSRSD 60
61 ELKRHMRTHT GQSQRRLKKA SVQKQEFLTV SGIPTIASGV MIHQPIPQVL PANMAINVQA 120
121 VNGGNIIHAP NAVHPMVIPI MAQPAPIHAS AASFQPATSP MPISTYTPVP SQSFTSFQSS 180
181 IGSIQSNSDV SSIFSNMNVR VNTPRSVPNS PNDGYLHQQH IPQQYQHQTA SPSVAKQQKT 240
241 FAHSLASALS TLQKRTPVSA PSTTIESPSS PSDSSHTSAS SSAISLPFSN APSQLAVAKE 300
301 LESVYLDSNR YTTKTRRERA KFEIPEEQEE DTNNSSSGSN EEEHESLDHE SSKSRKKLSG 360
361 VKLPPVRNLL KQIDVFNGPK RV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
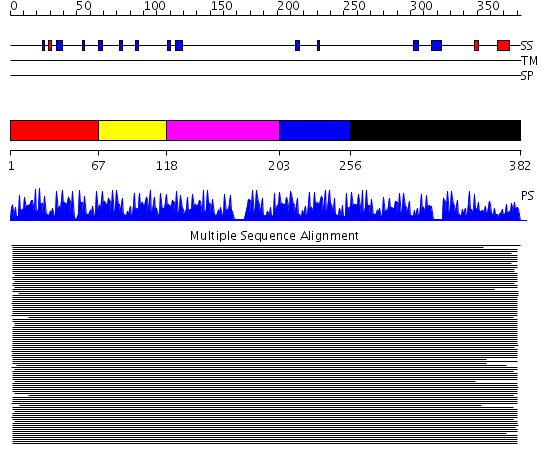
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..66] | 103.927757 | Five-finger GLI1 | |
| 2 | View Details | [67..117] | 103.927757 | Five-finger GLI1 | |
| 3 | View Details | [118..202] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [203..255] | 128.68867 | No description for 1lu6A_ was found. | |
| 5 | View Details | [256..382] | 128.68867 | No description for 1lu6A_ was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)