













| General Information: |
|
| Name(s) found: |
JHD1 /
YER051W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
histone demethylation
[IDA |
| Molecular Function: |
histone demethylase activity (H3-K36 specific)
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQDPNICQHC QLKDNPGALI WVKCDSCPQW VHVKCVPLKR IHYSNLTSSE VLSYPNSAKQ 60
61 IKSYRCPNHK EGEYLTAYAL ITQKGKRQRN KENPEDSHIN KRYNFRKKKL LDYIALNEGE 120
121 SKRDKMNHPH KESFMKSFEK WKNGSNIINA ADFAEKFDNI DVPYKIIDPL NSGVYVPNVG 180
181 TDNGCLTVNY ITEMIGEDYH VDVMDVQSQM NENWNLGSWN EYFTNTEPDR RDRIRNVISL 240
241 EVSNIEGLEL ERPTAVRQND LVDKIWSFNG HLEKVNGEKA EENDPKPKVT KYILMSVKDA 300
301 YTDFHLDFAG TSVYYNVISG QKKFLLFPPT QSNIDKYIEW SLKEDQNSVF LGDILEDGIA 360
361 MELDAGDLFM IPAGYIHAVY TPVDSLVFGG NFLTIRDLET HLKIVEIEKL TKVPRRFTFP 420
421 KFDQVMGKLC EYLALDKNKI TSDVSDGDLL SRTTNCAIQS LHAYVIKPEV KYKPLNFTSK 480
481 KHLAKALADL IS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
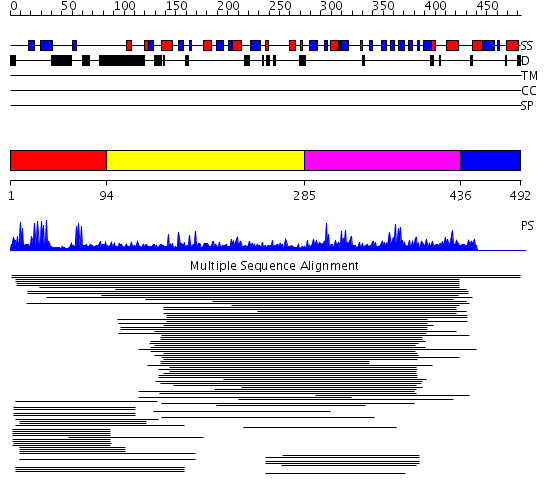
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | 5.036212 | PHD-finger No confident structure predictions are available. | |
| 2 | View Details | [94..284] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [285..435] | 2.958607 | JmjC domain No confident structure predictions are available. | |
| 4 | View Details | [436..492] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)