













| General Information: |
|
| Name(s) found: |
URA3 /
YEL021W
[SGD]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 267 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[ISS |
| Biological Process: |
'de novo' pyrimidine base biosynthetic process
[TAS pyrimidine base biosynthetic process [TAS |
| Molecular Function: |
orotidine-5'-phosphate decarboxylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKATYKERA ATHPSPVAAK LFNIMHEKQT NLCASLDVRT TKELLELVEA LGPKICLLKT 60
61 HVDILTDFSM EGTVKPLKAL SAKYNFLLFE DRKFADIGNT VKLQYSAGVY RIAEWADITN 120
121 AHGVVGPGIV SGLKQAAEEV TKEPRGLLML AELSCKGSLA TGEYTKGTVD IAKSDKDFVI 180
181 GFIAQRDMGG RDEGYDWLIM TPGVGLDDKG DALGQQYRTV DDVVSTGSDI IIVGRGLFAK 240
241 GRDAKVEGER YRKAGWEAYL RRCGQQN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
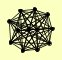
|
NAS6 RPN1 RPN2 RPN3 RPN5 RPN6 RPN7 RPN8 RPN9 RPT5 UBP6 URA3 |
| View Details | Krogan NJ, et al. (2006) |
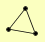
|
FOL1 URA3 YMR074C |
| View Details | Ho Y, et al. (2002) |

|
RNQ1 URA3 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
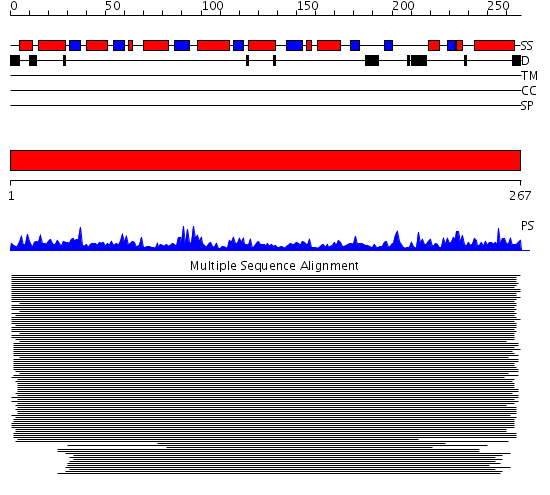
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 1607.522879 | Orotidine 5'-monophosphate decarboxylase (OMP decarboxylase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)