













| General Information: |
|
| Name(s) found: |
PPM1 /
YDR435C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
C-terminal protein amino acid methylation
[IDA |
| Molecular Function: |
C-terminal protein carboxyl methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERIIQQTDY DALSCKLAAI SVGYLPSSGL QRLSVDLSKK YTEWHRSYLI TLKKFSRRAF 60
61 GKVDKAMRSS FPVMNYGTYL RTVGIDAAIL EFLVANEKVQ VVNLGCGSDL RMLPLLQMFP 120
121 HLAYVDIDYN ESVELKNSIL RESEILRISL GLSKEDTAKS PFLIDQGRYK LAACDLNDIT 180
181 ETTRLLDVCT KREIPTIVIS ECLLCYMHNN ESQLLINTIM SKFSHGLWIS YDPIGGSQPN 240
241 DRFGAIMQSN LKESRNLEMP TLMTYNSKEK YASRWSAAPN VIVNDMWEIF NAQIPESERK 300
301 RLRSLQFLDE LEELKVMQTH YILMKAQW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
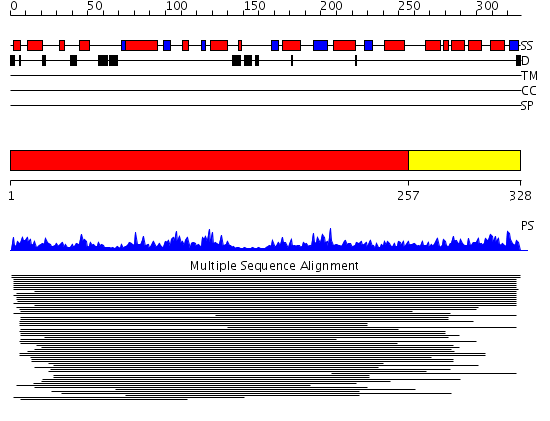
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..256] | 13.048977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [257..328] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)