













| General Information: |
|
| Name(s) found: |
FBpp0300193
[FlyBase]
RSG7-PA / FBpp0074124 [FlyBase] |
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
heterotrimeric G-protein complex
[IEA]
|
| Biological Process: |
intracellular signaling pathway
[IEA]
regulation of G-protein coupled receptor protein signaling pathway [ISS] |
| Molecular Function: |
signal transducer activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTMNSEADK TQATNASSTA APAKTDCTSS NSSDPLAVAI PEPQSATAVV GGGAGGAAPG 60
61 TATPSPTPLA NGSNNGTGSS VSPAATGTGA VATAASAPAA AGTTSASPPA SNPTTASTST 120
121 ASVVGAKTAA ATTTSSSSAT SSSTAAAGSK QSGSSGGLLT VSGNHHHHHH AHPQQSAAGQ 180
181 SAAQPPQPPT ATALAVPSAS HHQRGGKNED APNILVYKKM EAIIEKMQAE STGVAVRTVK 240
241 AFMSKVPSVF TGADLVAWIL KNFDVEDVTE ALHFAHLLSS HGYIFPIDDH ALTVKNDGTF 300
301 YRFQTPYFWP SNCWEPENTD YAVYLCKRTM QNKTRLELAD YEAENLAKLQ KMFSRKWEFI 360
361 FMQAESQSKV AKKRDKLERK VLDSQERAFW DVHRPMPGCV NTTEIDIKKA YRRGGSSHGT 420
421 GSAGASLAKN PVEQLTRIIA LRKQKLERRT IKVSKAAEAL VAYYDQYNEF DYFITSPELP 480
481 NPWQTDSTEM WDTEKNSKEV PVRRVKRWAF SLRELLNDAI GREQFTKFLE KEYSGENLKF 540
541 WESVQEMKAL PQSEIKEAIQ KIWQEFLAPE APCPVNVDSK SVELAREAVN SPSGPNRWCF 600
601 DVAASHVYHL MKSDSYSRYL RSDMYKDYLN CSRKKIKSIP NLFGVKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
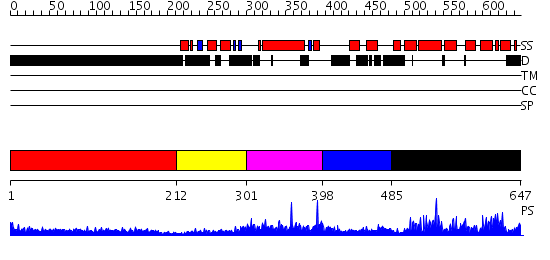
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..211] | 10.202996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [212..300] | 32.113509 | No description for PF00610.12 was found. No confident structure predictions are available. | |
| 3 | View Details | [301..397] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [398..484] | 1.53 | The Crystal Structure of a Biologically Active Peptide (SIGK) Bound to a G Protein Beta:Gamma Heterodimer | |
| 5 | View Details | [485..647] | 56.154902 | Structure of the regulator of G-protein signaling domain of RGS6 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)