













| General Information: |
|
| Name(s) found: |
pqe-1 /
CE39256
[WormBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 672 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: | NONE FOUND |
| Molecular Function: |
exonuclease activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFNGGYGSGN SFNLQNYAPI DPMTGIPGFG PSQNAQQQQQ QASAPGTSSG GPSQAVSGAS 60
61 SGASMKTEPV ARQVSTAQMK RDLEQAAVYV PTPIPKKDAK SSENRAKEKQ KPMNFQKRPT 120
121 SSASVDSNDD GVHIPAKRMA HASSVPGPSR SKPPMIGAVK NRPNHTEMLD KRNKESEEKR 180
181 RKDRDELERL RNKKHTTEEE KIKMARLQNA LKVVGKAAGL KATVKKELTG SPAKKQKPAP 240
241 AVPKILDFSV GRTFTAIRQT AIKLVFDTFL ERDSPNAARE AQEFELSIAK QYTDGQKYRI 300
301 NIGHKVAALR KENTSGILEV NKNAVSHDKI LAGGPKDNCT VARGRKTHVD HRQLSIEKLH 360
361 PLLLQFKLTT SELETNAYPM RRDGSTKAVS IADTVYTQNK KMFLDDYDMS RNCSRCNKEF 420
421 KLSPNGTMIR STGICRYHNR GVAINGKRDT FRKRYSCCNE EFNVALGCKF SDVHVTDQLF 480
481 KKELSTFVST PVPVPNDQRS TRVYALDCEM VYTIAGPALA RLTMVDMQRN RVLDVFVKPP 540
541 TDVLDPNTEF SGLTMEQINS APDTLKTCHQ KLFKYVNADT ILIGHSLESD LKAMRVVHKN 600
601 VIDTAILFRS TRDTKVALKV LSAKLLHKNI QGDNEDAIGH DSMEDALTCV DLIFYGLRNP 660
661 ESIAIREANT NC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
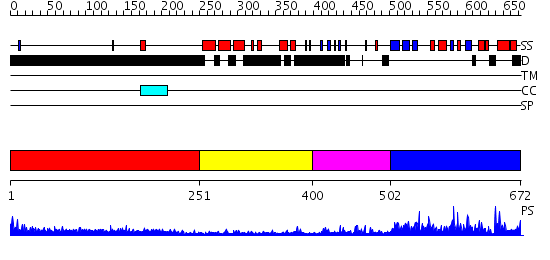
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | 1.17 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | |
| 2 | View Details | [251..399] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [400..501] | 1.0 | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | |
| 4 | View Details | [502..672] | 43.09691 | human ISG20 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)