













| General Information: |
|
| Name(s) found: |
ILVE_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 44 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 309 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTKKADYIW FNGEMVRWED AKVHVMSHAL HYGTSVFEGI RCYDSHKGPV VFRHREHMQR 60
61 LHDSAKIYRF PVSQSIDELM EACRDVIRKN NLTSAYIRPL IFVGDVGMGV NPPAGYSTDV 120
121 IIAAFPWGAY LGAEALEQGI DAMVSSWNRA APNTIPTAAK AGGNYLSSLL VGSEARRHGY 180
181 QEGIALDVNG YISEGAGENL FEVKDGVLFT PPFTSSALPG ITRDAIIKLA KELGIEVREQ 240
241 VLSRESLYLA DEVFMSGTAA EITPVRSVDG IQVGEGRCGP VTKRIQQAFF GLFTGETEDK 300
301 WGWLDQVNQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
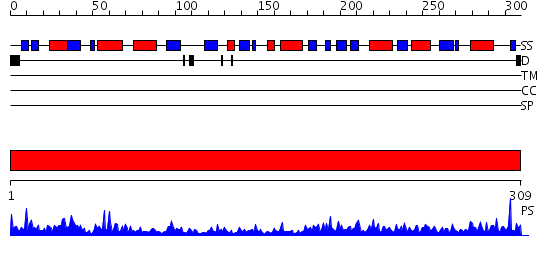
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..309] | 102.0 | Branched-chain aminoacid aminotransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)