













| General Information: |
|
| Name(s) found: |
FBpp0306732
[FlyBase]
gfzf-PB / FBpp0290855 [FlyBase] |
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1045 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[ISS]
glutathione binding [IDA] glutathione transferase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METPVEEHGM DLTKVSVVRS AKGVDMLCVD DYLYHFDRIG RNNTYRWLCN RRKDKSTPCK 60
61 SRICTMLPDP LDKTRHVVVD VVLAHIHPRC SDHEVNKVAQ RKRLSVMMMN DAYSTTQPKR 120
121 ARLYERANAG MNPPEFIYGS IDTSTVEASF QLDFEDKDRV YMLRRRDGRV MVCIDGHLYY 180
181 LAGKSYKGDI FRWTCVRKRD IECTARICTE ATLAGAHRLH AVDTDAHIHP HYSAAKLMHM 240
241 FSKHQIVLVE DENTSDVLAE CPNLEYFDPE NTFDTAQRGD DLESIIIEEC EDNYDIYMRT 300
301 DGSQGGGIQG SEELESQEGD DESSLVDPKM HHSSKWPAAF DVIANYLPPP SHNPLTETDL 360
361 TKFTFLPSAK GRKVLCLSQH LFHFDSQSRS NGHMFFTCIN RRDKANTCHV RITLDPVENG 420
421 PVVLRINGEH THNPDIKEIA RRLKQQAKLD QDNVHKLQKI EHSEEISFES DEGSASQGGD 480
481 TFEAENSVDE SYGHIDTRNV VMKKVISIDG AIKMEPESKA NMIVQYLKDG ANNISAKIEI 540
541 LPDALDELDP HSHTAYSSTP LPKVRQPQIT NIRRRRDVAE PAKGAQPDMD LSKICTLRSA 600
601 KGHELLCVDG YIYHAKNRGV ISRNYWVCIK SRDPDINCKS RISTATQKDG SIRVLRVYNS 660
661 HSHPFSEDDI KRRLYNEINK KNNKNLKFRP LHFIGKSLDQ IQQEYGDLAI ERLNVSNISS 720
721 DIVVHKKPRV SGSNNSRSAS LSVSHRADSH HEEDDVVIVE EEYQEQEEGV EMTMEGDQQY 780
781 MDVDNTDGII EVVEEVADEM ESYGSIVPIY TMKLYAVSDG PPSLAVRMTL KALDIQYQLI 840
841 NVDFCAMEHR SEEYSKMNPQ KEIPVLDDDG FYLSESIAIM QYLCDKYAPD STLYPQDVNV 900
901 RAVINQRLCF NMGFYYAPIS AHSMAPIFFD YKRTPMSLKK VQNALDVFET YLQRLGTKYA 960
961 AGENITIADF ALISATICLE AINFDLHQFT LVNKWYETFK VEYPQLWEIA NSGMQEISAF1020
1021 EQNPPDMSHM EHPFHPTRKS MGLKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
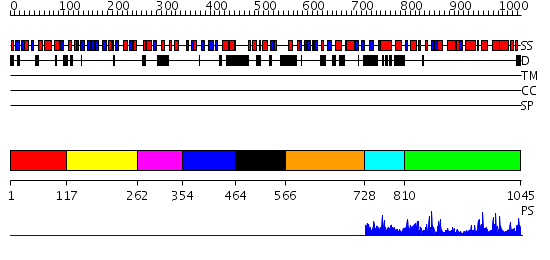
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 7.102373 | No description for PF04500.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [117..261] | 4.09691 | No description for PF04500.7 was found. No confident structure predictions are available. | |
| 3 | View Details | [262..353] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [354..463] | 32.075721 | No description for PF04500.7 was found. No confident structure predictions are available. | |
| 5 | View Details | [464..565] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [566..727] | 32.79588 | No description for PF04500.7 was found. No confident structure predictions are available. | |
| 7 | View Details | [728..809] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [810..1045] | 49.045757 | Crystal structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)