













| General Information: |
|
| Name(s) found: |
NARZ_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 1246 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKLLDRFRY FKQKGETFAD GHGQVMHSNR DWEDSYRQRW QFDKIVRSTH GVNCTGSCSW 60
61 KIYVKNGLVT WEIQQTDYPR TRPDLPNHEP RGCPRGASYS WYLYSANRLK YPLIRKRLIE 120
121 LWREALKQHS DPVLAWASIM NDPQKCLSYK QVRGRGGFIR SNWQELNQLI AAANVWTIKT 180
181 YGPDRVAGFS PIPAMSMVSY AAGTRYLSLL GGTCLSFYDW YCDLPPASPM TWGEQTDVPE 240
241 SADWYNSSYI IAWGSNVPQT RTPDAHFFTE VRYKGTKTIA ITPDYSEVAK LCDQWLAPKQ 300
301 GTDSALAMAM GHVILKEFHL DNPSDYFINY CRRYSDMPML VMLEPRDDGS YVPGRMIRAS 360
361 DLVDGLGESN NPQWKTVAVN TAGELVVPNG SIGFRWGEKG KWNLESIAAG TETELSLTLL 420
421 GQHDAVAGVA FPYFGGIENP HFRSVKHNPV LVRQLPVKNL TLVDGNTCPV VSVYDLVLAN 480
481 YGLDRGLEDE NSAKDYAEIK PYTPAWGEQI TGVPRQYIET IAREFADTAH KTHGRSMIIL 540
541 GAGVNHWYHM DMNYRGMINM LIFCGCVGQS GGGWAHYVGQ EKLRPQTGWL PLAFALDWNR 600
601 PPRQMNSTSF FYNHSSQWRY EKVSAQELLS PLADASKYSG HLIDFNVRAE RMGWLPSAPQ 660
661 LGRNPLGIKA EADKAGLSPT EFTAQALKSG DLRMACEQPD SSSNHPRNLF VWRSNLLGSS 720
721 GKGHEYMQKY LLGTESGIQG EELGASDGIK PEEVEWQTAA IEGKLDLLVT LDFRMSSTCL 780
781 FSDIVLPTAT WYEKDDMNTS DMHPFIHPLS AAVDPAWESR SDWEIYKGIA KAFSQVCVGH 840
841 LGKETDVVLQ PLLHDSPAEL SQPCEVLDWR KGECDLIPGK TAPNIVAVER DYPATYERFT 900
901 SLGPLMDKLG NGGKGISWNT QDEIDFLGKL NYTKRDGPAQ GRPLIDTAID ASEVILALAP 960
961 ETNGHVAVKA WQALGEITGR EHTHLALHKE DEKIRFRDIQ AQPRKIISSP TWSGLESDHV1020
1021 SYNAGYTNVH ELIPWRTLSG RQQLYQDHPW MRAFGESLVA YRPPIDTRSV SEMRQIPPNG1080
1081 FPEKALNFLT PHQKWGIHST YSENLLMLTL SRGGPIVWIS ETDARELTIV DNDWVEVFNA1140
1141 NGALTARAVV SQRVPPGMTM MYHAQERIMN IPGSEVTGMR GGIHNSVTRV CPKPTHMIGG1200
1201 YAQLAWGFNY YGTVGSNRDE FIMIRKMKNV NWLDDEGRDQ VQEAKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
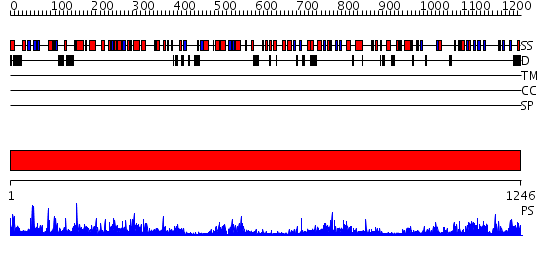
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1246] | 1000.0 | Crystal structure of Nitrate Reductase A, NarGHI, from Escherichia coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)