













| General Information: |
|
| Name(s) found: |
DPO3_STRPN
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Streptococcus pneumoniae TIGR4 |
| Length: | 1463 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSFEILMN QLGMPAEMRQ APALAQANIE RVVVHKISKV WEFHFVFSNI LPIEIFLELK 60
61 KGLSEEFSKT GNKAVFEIKA RSQEFSNQLL QSYYREAFSE GPCASQGFKS LYQNLQVRAE 120
121 GNQLFIEGSE AIDKEHFKKN HLPNLAKQLE KFGFPTFNCQ VEKNDVLTQE QEEAFHAENE 180
181 QIVQAANEEA LRAMEQLEQM APPPAEEKPA FDFQAKKAAA KPKLDKAEIT PMIEVTTEEN 240
241 RLVFEGVVFD VEQKVTRTGR VLINFKMTDY TSSFSMQKWV KNEEEAQKFD LIKKNSWLRV 300
301 RGNVEMNNFT RDLTMNVQDL QEVVHYERKD LMPEGERRVE FHAHTNMSTM DALPEVEEIV 360
361 ATAAKWGHKA VAITDHGNVQ SFPHGYKAAK KAGIQLIYGM EANIVEDRVP IVYNEVEMDL 420
421 SEATYVVFDV ETTGLSAIYN DLIQVAASKM YKGNVIAEFD EFINPGHPLS AFTTELTGIT 480
481 DDHVKNAKPL EQVLQEFQEF CKDTVLVAHN ATFDVGFMNA NYERHDLPKI SQPVIDTLEF 540
541 ARNLYPEYKR HGLGPLTKRF GVALEHHHMA NYDAEATGRL LFIFIKEVAE KHGVTDLARL 600
601 NIDLISPDSY KKARIKHATI YVKNQVGLKN IFKLVSLSNT KYFEGVPRIP RTVLDAHREG 660
661 LILGSACSEG EVFDVVVSQG VDAAVEVAKY YDFIEVMPPA IYAPLIAKEQ VKDMEELQTI 720
721 IKSLIEVGDR LGKPVLATGN VHYIEPEEEI YREIIVRSLG QGAMINRTIG HGEHAQPAPL 780
781 PKAHFRTTNE MLDEFAFLGE ELARKLVIEN TNALAEIFES VEVVKGDLYT PFIDKAEETV 840
841 AELTYKKAFE IYGNPLPDIV DLRIEKELTS ILGNGFAVIY LASQMLVQRS NERGYLVGSR 900
901 GSVGSSFVAT MIGITEVNPL SPHYVCGQCQ YSEFITDGSY GSGFDMPHKD CPNCGHKLSK 960
961 NGQDIPFETF LGFDGDKVPD IDLNFSGEDQ PSAHLDVRDI FGEEYAFRAG TVGTVAAKTA1020
1021 YGFVKGYERD YGKFYRDAEV ERLAQGAAGV KRTTGQHPGG IVVIPNYMDV YDFTPVQYPA1080
1081 DDVTAEWQTT HFNFHDIDEN VLKLDVLGHD DPTMIRKLQD LSGIDPNKIP MDDEGVMALF1140
1141 SGTDVLGVTP EQIGTPTGML GIPEFGTNFV RGMVDETHPT TFAELLQLSG LSHGTDVWLG1200
1201 NAQDLIKQGI ADLSTVIGCR DDIMVYLMHA GLEPKMAFTI MERVRKGLWL KISEEERNGY1260
1261 IEAMKANKVP EWYIESCGKI KYMFPKAHAA AYVMMALRVA YFKVHHPIYY YCAYFSIRAK1320
1321 AFDIKTMGAG LEVIKRRMEE ISEKRKNNEA SNVEIDLYTT LEIVNEMWER GFKFGKLDLY1380
1381 CSQATEFLID GDTLIPPFVA MDGLGENVAK QLVRAREEGE FLSKTELRKR GGLSSTLVEK1440
1441 MDEMGILGNM PEDNQLSLFD ELF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
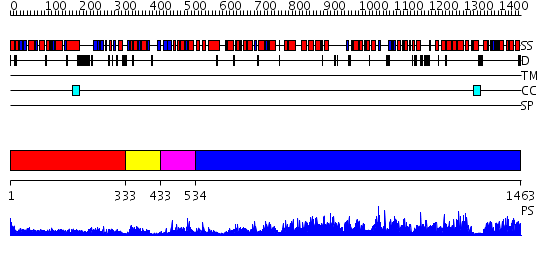
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..332] | 4.30103 | No description for 2fkjA was found. | |
| 2 | View Details | [333..432] | 3.05 | Hypothetical protein YcdX | |
| 3 | View Details | [433..533] | 26.39794 | No description for 2guiA was found. | |
| 4 | View Details | [534..1463] | 1000.0 | No description for 2hnhA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)