













| General Information: |
|
| Name(s) found: |
SENP2
[HGNC (HUGO)]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYRWLVRILG TIFRFCDRSV PPARALLKRR RSDSTLFSTV DTDEIPAKRP RLDCFIHQVK 60
61 NSLYNAASLF GFPFQLTTKP MVTSACNGTR NVAPSGEVFS NSSSCELTGS GSWNNMLKLG 120
121 NKSPNGISDY PKIRVTVTRD QPRRVLPSFG FTLNSEGCNR RPGGRRHSKG NPESSLMWKP 180
181 QEQAVTEMIS EESGKGLRRP HCTVEEGVQK EEREKYRKLL ERLKESGHGN SVCPVTSNYH 240
241 SSQRSQMDTL KTKGWGEEQN HGVKTTQFVP KQYRLVETRG PLCSLRSEKR CSKGKITDTE 300
301 TMVGIRFENE SRRGYQLEPD LSEEVSARLR LGSGSNGLLR RKVSIIETKE KNCSGKERDR 360
361 RTDDLLELTE DMEKEISNAL GHGPQDEILS SAFKLRITRG DIQTLKNYHW LNDEVINFYM 420
421 NLLVERNKKQ GYPALHVFST FFYPKLKSGG YQAVKRWTKG VNLFEQEIIL VPIHRKVHWS 480
481 LVVIDLRKKC LKYLDSMGQK GHRICEILLQ YLQDESKTKR NSDLNLLEWT HHSMKPHEIP 540
541 QQLNGSDCGM FTCKYADYIS RDKPITFTQH QMPLFRKKMV WEILHQQLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
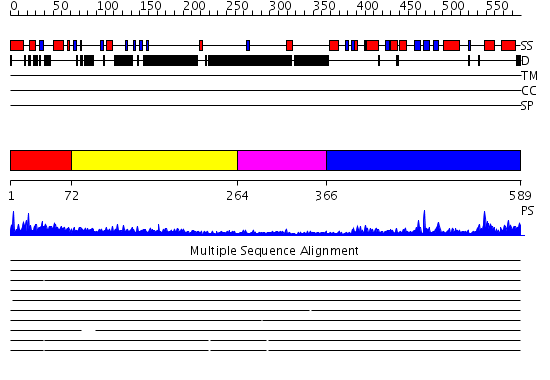
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [72..263] | 3.335994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [264..365] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [366..589] | 65.69897 | Structure of human Senp2 in complex with SUMO-1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)