













| General Information: |
|
| Name(s) found: |
gi|39995196
[NCBI NR]
gi|39981958 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Geobacter sulfurreducens PCA |
| Length: | 280 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
disulfide oxidoreductase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCDHKNIYLP HLATIEEIVD ETPDIRTFRL VFQDERVREH FTFRAGQFAE YSAFGAGEAT 60
61 FCIASAPTRQ GYIECCFRAV GRVTEALRSL ETGDTIGVRG PYGNSFPVEE FFGKNLVFVA 120
121 GGIALPPLRT LIWQCLDWRE KFGDITIVYG ARTEADLVYK RELREWEERS DVRLVKTVDP 180
181 GGNSPSWDGQ VGFVPTVLEQ AAPAADNTIA LVCGPPVMIK FTLPVLEKLG FADTAIYTTL 240
241 ENRMKCGLGK CGRCNVGNVY VCKDGPVFTA AQVKAMPQEF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
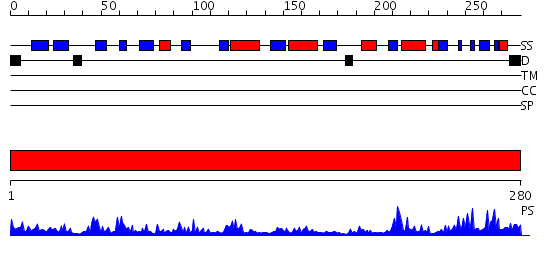
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..280] | 49.39794 | cytochrome b5 reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)