













| General Information: |
|
| Name(s) found: |
PIN4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 668 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METSSFENAP PAAINDAQDN NINTETNDQE TNQQSIETRD AIDKENGVQT ETGENSAKNA 60
61 EQNVSSTNLN NAPTNGALDD DVIPNAIVIK NIPFAIKKEQ LLDIIEEMDL PLPYAFNYHF 120
121 DNGIFRGLAF ANFTTPEETT QVITSLNGKE ISGRKLKVEY KKMLPQAERE RIEREKREKR 180
181 GQLEEQHRSS SNLSLDSLSK MSGSGNNNTS NNQLFSTLMN GINANSMMNS PMNNTINNNS 240
241 SNNNNSGNII LNQPSLSAQH TSSSLYQTNV NNQAQMSTER FYAPLPSTST LPLPPQQLDF 300
301 NDPDTLEIYS QLLLFKDREK YYYELAYPMG ISASHKRIIN VLCSYLGLVE VYDPRFIIIR 360
361 RKILDHANLQ SHLQQQGQMT SAHPLQPNST GGSMNRSQSY TSLLQAHAAA AANSISNQAV 420
421 NNSSNSNTIN SNNGNGNNVI INNNSASSTP KISSQGQFSM QPTLTSPKMN IHHSSQYNSA 480
481 DQPQQPQPQT QQNVQSAAQQ QQSFLRQQAT LTPSSRIPSG YSANHYQINS VNPLLRNSQI 540
541 SPPNSQIPIN SQTLSQAQPP AQSQTQQRVP VAYQNASLSS QQLYNLNGPS SANSQSQLLP 600
601 QHTNGSVHSN FSYQSYHDES MLSAHNLNSA DLIYKSLSHS GLDDGLEQGL NRSLSGLDLQ 660
661 NQNKKNLW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
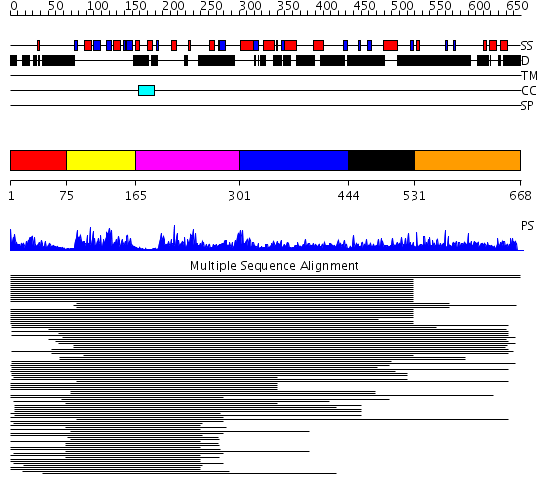
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | 47.221849 | Poly(A)-binding protein | |
| 2 | View Details | [75..164] | 57.0 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 3 | View Details | [165..300] | 57.0 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 4 | View Details | [301..443] | 2.206995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [444..530] | 8.522879 | poly(A) binding protein | |
| 6 | View Details | [531..668] | 2.154996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)