













| General Information: |
|
| Name(s) found: |
IOC2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 812 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRTKRTRGTR NVGASMPAGA ANADSEDWKE YVSEDIIAQL NKHQLPYSEI LDEKIADLAN 60
61 HWHFQYVMAW LSNVCESYTT TTFNTDQYGG SSTKCLWKNI KFDEGVFVTD VFSKIDGKDS 120
121 NYYNDEVDVD EGSQNLYDRI RLQLLHQLAG NKSGQLKDWN VIVNHHLQNS SAYSDLVTDL 180
181 PFLELEIARQ FDIIYSIIKL IEMKNMIFKN YLANNLHLFT FSEVILDDDN SGGEEMKSLF 240
241 ALPNVGVLVK KTIHRVKEGS SSQVSQTLNI PIKLQNCTIK ESDPDIPDSV ELIHLEYSHD 300
301 IDAYLQSITI DYDVITTNWG SMLEYWSENK SSKAIDEFIT NLIPVYAEHR LYSAKLLANR 360
361 EKERAIAELM TRRKRSSRLV AKEEENKKKD LESEWFEKLD EREQFIRHRN KLVSKEIKKI 420
421 KDLLWNQLWQ LYDQDYRDEK LTRRNELKDR SGSGTPFFET SLGREEDNPL NEIDIGVLDH 480
481 GPNFQSSIIP VEPPIPGTVG PLQTGDVPEL PSDFCITKEE LDELANYGIF TPQQEPDNQD 540
541 SVFQCPGEPE LAPMIITEDT ETDLFNNRPL ICCDHCYRWQ HWECQPPKII ELISSTTKSP 600
601 QHTLSQRDFG VIIMGNSHGN RRSSRRPQST LEPSTKSSRP TDKRKPLSEC STFICAWCIR 660
661 DLELELRNIF VPELKIIRAK QRKQQEDRER RKKMKEEKKR LEELAKKREL TESVSPPVFN 720
721 NAFANMSSST TPSIAAYEKT NPAINPAPNV NAAHPIITYS QQTGSKTVPQ APQAPQTSQA 780
781 SIQPQQQQQQ QQQQQPLHPK EQNFHFQFPP TN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
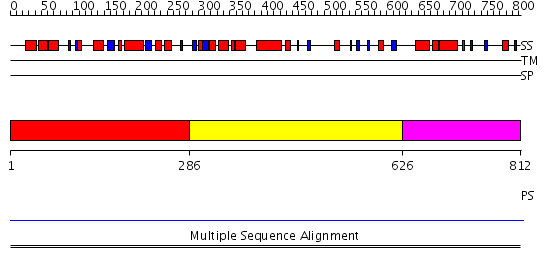
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..285] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [286..625] | 7.45 | Microbial transglutaminase | |
| 3 | View Details | [626..812] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|