













| General Information: |
|
| Name(s) found: |
ECM32_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1121 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFQCRTCSQ AYDAEQMMKH LSSTRHKTVF DTSNDEDICC EECQDKNIHQ LQIIRFGGED 60
61 MVLLCNSCFR KEYSETERPS TSYSLQNGSI LKFWEKYVKV RECCCDECGE ESNLNANRNG 120
121 EVLCDKCLPK SNRAKDFVSE KSGRFLYIYL GLNETQNSTR KPRKKGGRRV GRGKKGRKGA 180
181 KIKKEKKETF EAKISRIAYE VKKENSTIQS SSSSNLRNFK GFKAVESDPV VAAKVSKSET 240
241 SRSNPGPSNR NKGKGNKANH KKNSGNGIGK EKERKTNIRN NVRNSQPIPE DRKNTNSHVT 300
301 TNSGGKGKNE SVDKHQLPQP KALNGNGSGS TNTTGLKKGK KDHAGQKTKG NDKTGNKNPR 360
361 EAKLNSAGRK NALGKKSNNQ PNKGTSRWTI GSDTESSREP SISPNENTTS ITKSRNRNKK 420
421 ASKPTLNEKS KTTTMPKKLE TKNQEKNNGK TKDGKLIYEE GEPLTRYNTF KSTLSYPDLN 480
481 TYLNDYSFAL FLEQKLENEF VQNFNILWPR NEKDTAFIIN VEKNNNSELE KLLPANLLAL 540
541 GRPAFNERQP FFFCTQDEQK VWYIFIKELS IQRGKYVLLV ELFSWNNLSL PTKNGSSQFK 600
601 LLPTSAQTSR ILFAMTRITN PKFIDLLLGQ KPIKEIYFDN RLKFSSDKLN RSQKTAVEHV 660
661 LNNSITILQG PPGTGKTSTI EEIIIQVIER FHAFPILCVA ASNIAIDNIA EKIMENRPQI 720
721 KILRILSKKK EQQYSDDHPL GEICLHNIVY KNLSPDMQVV ANKTRRGEMI SKSEDTKFYK 780
781 EKNRVTNKVV SQSQIIFTTN IAAGGRELKV IKECPVVIMD EATQSSEAST LVPLSLPGIR 840
841 NFVFVGDEKQ LSSFSNIPQL ETSLFERVLS NGTYKNPLML DTQYRMHPKI SEFPIKKIYN 900
901 GELKDGVTDE QKAWPGVQHP LFFYQCDLGP ESRVRSTQRD IVGFTYENKH ECVEIVKIIQ 960
961 ILMLDKKVPL EEIGVITPYS AQRDLLSDIL TKNVVINPKQ ISMQQEYDEI ELFNAAGSQG1020
1021 TAGSLQNNVI NIINGLHVAT VDSFQGHEKS FIIFSCVRNN TENKIGFLRD KRRLNVALTR1080
1081 AKHGLIVVGN KNVLRKGDPL WKDYITYLEE QEVIFTDLTA Y |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
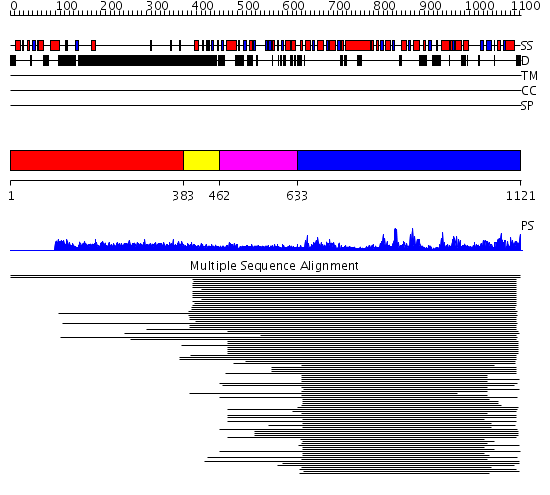
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..382] | 8.07 | RBP1 | |
| 2 | View Details | [383..461] | 10.24 | No description for 1l8mA was found. | |
| 3 | View Details | [462..632] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [633..1121] | 80.30103 | DEXX box DNA helicase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)