













| General Information: |
|
| Name(s) found: |
HRQ1_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1077 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEGPIKKKL KSAGQGSGKT DAFRNFEQFF FRLNTLYTFL ICRKHVVPTF KTLCGPIETA 60
61 LKRTVTKEDL AMVMALMPRE CVFKYIDENQ IYTETKIFDF NNGGFQQKEN DIFELKDVDD 120
121 QNQTQKSTQL LIFEFIDGTM QRSWSASDRF SQIKIPTYTT EEMKKMISKR EALFKSRLRE 180
181 FILEKEKANL DPFSELTNLA QKYIPRERDY EDPIEAMMKA KQESNEMSIP NYSNNSVITT 240
241 IPQMIEKLKS TEFYASQIKH CFTIPSRTAK YKGLCFELAP EVYQGMEHEN FYSHQADAIN 300
301 SLHQGENVII TTSTSSGKSL IYQLAAIDLL LKDPESTFMY IFPTKALAQD QKRAFKVILS 360
361 KIPELKNAVV DTYDGDTEPE ERAYIRKNAR VIFTNPDMIH TSILPNHANW RHFLYHLKLV 420
421 VVDELHIYKG LFGSHVALVM RRLLRLCHCF YENSGLQFIS CSATLKSPVQ HMKDMFGINE 480
481 VTLIHEDGSP TGAKHLVVWN PPILPQHERK RENFIRESAK ILVQLILNNV RTIAFCYVRR 540
541 VCELLMKEVR NIFIETGRED LVTEVMSYRG GYSASDRRKI EREMFHGNLK AVISTNALEL 600
601 GIDIGGLDAV LMCGFPLSMA NFHQQSGRAG RRNNDSLTLV VASDSPVDQH YVAHPESLLE 660
661 VNNFESYQDL VLDFNNILIL EGHIQCAAFE LPINFERDKQ YFTESHLRKI CVERLHHNQD 720
721 GYHASNRFLP WPSKCVSLRG GEEDQFAVVD ITNGRNIIIE EIEASRTSFT LYDGGIFIHQ 780
781 GYPYLVKEFN PDERYAKVQR VDVDWVTNQR DFTDVDPQEI ELIRSLRNSD VPVYFGKIKT 840
841 TIIVFGFFKV DKYKRIIDAI ETHNPPVIIN SKGLWIDMPK YALEICQKKQ LNVAGAIHGA 900
901 QHAIMGMLPR FIVAGVDEIQ TECKAPEKEF AERQTKRKRP ARLIFYDSKG GKYGSGLCVK 960
961 AFEHIDDIIE SSLRRIEECP CSDGCPDCVA ASFCKENSLV LSKPGAQVVL HCILGHSEDS1020
1021 FIDLIKDGPE PNMPEIKVET VIPVSEHVNF SDDFKIIDVR RATKDDTHTN EIIKKEI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
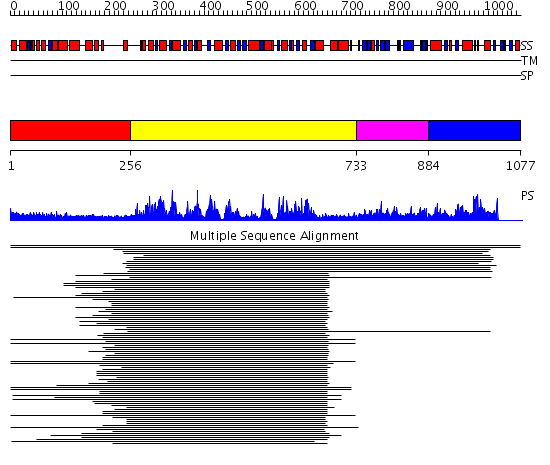
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..255] | 27.684997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [256..732] | 90.39794 | Initiation factor 4a | |
| 3 | View Details | [733..883] | 1.195988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [884..1077] | 1.022972 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [831..1015] | 36.136677 | No description for PF09369.1 was found. No confident structure predictions are available. | |
| 6 | View Details | [1016..1077] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)