













| General Information: |
|
| Name(s) found: |
KIN13_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 794 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi stack
[IDA]
plasma membrane [IDA] |
| Biological Process: |
trichome morphogenesis
[IMP]
|
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGQMQQNNA AAATALYDGA LPTNDAGDAV MARWLQSAGL QHLASPVAST GNDQRHLPNL 60
61 LMQGYGAQTA EEKQRLFQLM RNLNFNGEST SESYTPTAHT SAAMPSSEGF FSPEFRGDFG 120
121 AGLLDLHAMD DTELLSEHVI TEPFEPSPFM PSVNKEFEED YNLAANRQQR QQTEAEPLGL 180
181 LPKSDKENNS VAKIKVVVRK RPLNKKETAK KEEDVVTVSD NSLTVHEPRV KVDLTAYVEK 240
241 HEFCFDAVLD EDVSNDEVYR ATIEPIIPII FQRTKATCFA YGQTGSGKTF TMKPLPIRAV 300
301 EDLMRLLRQP VYSNQRFKLW LSYFEIYGGK LFDLLSERKK LCMREDGRQQ VCIVGLQEYE 360
361 VSDVQIVKDF IEKGNAERST GSTGANEESS RSHAILQLVV KKHVEVKDTR RRNNDSNELP 420
421 GKVVGKISFI DLAGSERGAD TTDNDRQTRI EGAEINKSLL ALKECIRALD NDQLHIPFRG 480
481 SKLTEVLRDS FVGNSRTVMI SCISPNAGSC EHTLNTLRYA DRVKSLSKSG NSKKDQTANS 540
541 MPPVNKDPLL GPNDVEDVFE PPQEVNVPET RRRVVEKDSN SSTSGIDFRQ PTNYREESGI 600
601 PSFSMDKGRS EPNSSFAGST SQRNNISSYP QETSDREEKV KKVSPPRGKG LREEKPDRPQ 660
661 NWSKRDVSSS DIPTLTNFRQ NASETASRQY ETASRQYETD PSLDENLDAL LEEEEALIAA 720
721 HRKEIEDTME IVREEMKLLA EVDQPGSMIE NYVTQLSFVL SRKAAGLVSL QARLARFQHR 780
781 LKEQEILSRK RVPR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
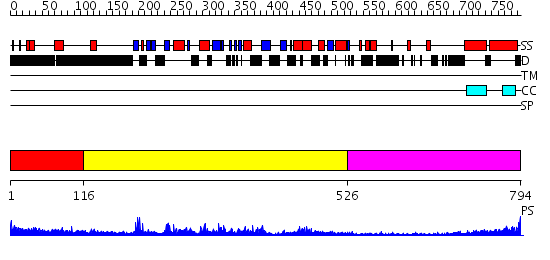
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 1.01 | Crystal Structure of the native Shank SAM domain. | |
| 2 | View Details | [116..525] | 72.221849 | No description for 2gryA was found. | |
| 3 | View Details | [526..794] | 11.154902 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)