













| General Information: |
|
| Name(s) found: |
AP2A2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1013 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
vesicle-mediated transport
[IEA]
protein transport [IEA] intracellular protein transport [IEA] |
| Molecular Function: |
protein binding
[IEA]
protein transporter activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGMRGLSVF ISDVRNCQNK EAERLRVDKE LGNIRTCFKN EKVLTPYKKK KYVWKMLYIH 60
61 MLGYDVDFGH MEAVSLISAP KYPEKQVGYI VTSCLLNENH DFLKLAINTV RNDIIGRNET 120
121 FQCLALTLVG NIGGRDFAES LAPDVQKLLI SSSCRPLVRK KAALCLLRLF RKNPDAVNVD 180
181 GWADRMAQLL DERDLGVLTS STSLLVALVS NNHEAYSSCL PKCVKILERL ARNQDVPQEY 240
241 TYYGIPSPWL QVKAMRALQY FPTIEDPSTR KALFEVLQRI LMGTDVVKNV NKNNASHAVL 300
301 FEALSLVMHL DAEKEMMSQC VALLGKFISV REPNIRYLGL ENMTRMLMVT DVQDIIKKHQ 360
361 SQIITSLKDP DISIRRRALD LLYGMCDVSN AKDIVEELLQ YLSTAEFSMR EELSLKAAIL 420
421 AEKFAPDLSW YVDVILQLID KAGDFVSDDI WFRVVQFVTN NEDLQPYAAS KAREYMDKIA 480
481 IHETMVKVSA YILGEYGHLL ARQPGCSASE LFSILHEKLP TVSTPTIPIL LSTYAKLLMH 540
541 AQPPDPELQK KVWAVFKKYE SCIDVEIQQR AVEYFELSKK GPAFMDVLAE MPKFPERQSS 600
601 LIKKAENVED TADQSAIKLR AQQQPSNAIV LADPQPVNGA PPPLKVPILS GSTDPESVAR 660
661 SLSHPNGTLS NIDPQTPSPD LLSDLLGPLA IEAPPGAVSY EQHGPVGAEG VPDEIDGSAI 720
721 VPVEEQTNTV ELIGNIAERF HALCLKDSGV LYEDPHIQIG IKAEWRGHHG RLVLFMGNKN 780
781 TSPLTSVQAL ILPPAHLRLD LSPVPDTIPP RAQVQSPLEV MNIRPSRDVA VLDFSYKFGT 840
841 NVVSAKLRIP ATLNKFLQPL QLTSEEFFPQ WRAISGPPLK LQEVVRGVRP LALPEMANLF 900
901 NSFHVTICPG LDPNPNNLVA STTFYSETTG AMLCLARIET DPADRTQLRL TVGSGDPTLT 960
961 FELKEFIKEQ LITIPMGSRA LVPAAGPAPS PAVQPPSPAA LADDPGAMLA GLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
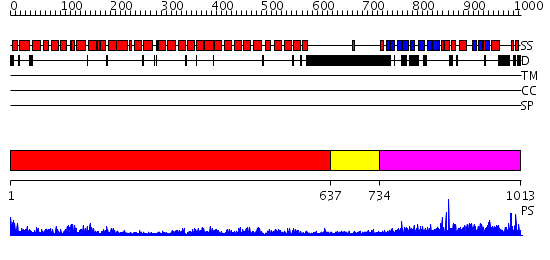
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..636] | 96.0 | Adaptin alpha C subunit N-terminal fragment | |
| 2 | View Details | [637..733] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [734..1013] | 78.522879 | Crystal structure of the alpha-adaptin appendage domain, from the AP2 adaptor complex, bound to 2 peptides from Synaptojanin170 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)