













| General Information: |
|
| Name(s) found: |
gi|190404894
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 463 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRATRTAA AKSLVKSKVA RNVMAASFVK RHASTSLFKQ ANKVESLGSI YLSGKKISVA 60
61 ANPFSITSNR FKSTSIEVPP MAESLTEGSL KEYTKNVGDF IKEDELLATI ETDKIDIEVN 120
121 SPVSGTVTKL NFKPEDTVTV GEELAQVEPG EAPAEGSGES KPEPTEQAEP SQGVAARENS 180
181 SEETASKKEA APKKEAAPKK EVTEPKKADQ PKKTVSKAQE PPVASNSFTP FPRTETRVKM 240
241 NRMRLRIAER LKESQNTAAS LTTFNEVDMS ALMEMRKLYK DEIIKKTGTK FGFMGLFSKA 300
301 CTLAAKDIPA VNGAIEGDQI VYRDYTDISV AVATPKGLVT PVVRNAESLS VLDIENEIVR 360
361 LSHKARDGKL TLEDMTGGTF TISNGGVFGS LYGTPIINSP QTAVLGLHGV KERPVTVNGQ 420
421 IVSRPMMYLA LTYDHRLLDG REAVTFLKTV KELIEDPRKM LLW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
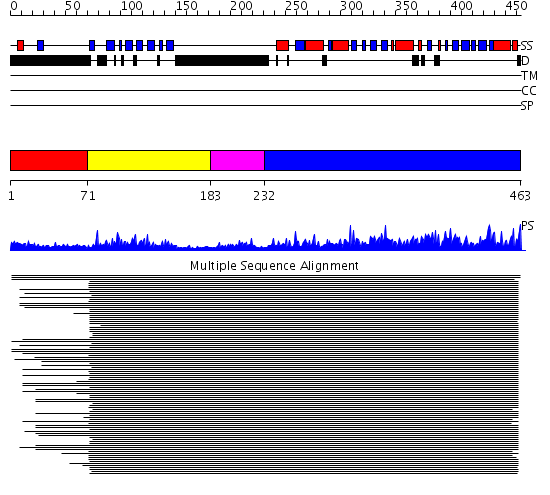
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..182] | 67.9897 | Ipoyl domain of dihydrolipoamide acetyltransferase | |
| 3 | View Details | [183..231] | 4.09691 | E3/E1 binding domain of dihydrolipoyl acetyltransferase | |
| 4 | View Details | [232..463] | 870.0 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)