













| General Information: |
|
| Name(s) found: |
gi|190408810
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 699 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTAQLQSEST SKIVLVTGGA GYIGSHTVVE LIENGYDCVV ADNLSNSTYD SVARLEVLTK 60
61 HHIPFYEVDL CDRKGLEKVF KEYKIDSVIH FAGLKAVGES TQIPLRYYHN NILGTVVLLE 120
121 LMQQYNVSKF VFSSSATVYG DATRFPNMIP IPEECPLGPT NPYGHTKYAI ENILNDLYNS 180
181 DKKSWKFAIL RYFNPIGAHP SGLIGEDPLG IPNNLLPYMA QVAVGRREKL YIFGDDYDSR 240
241 DGTPIRDYIH VVDLAKGHIA ALQYLEAYNE NEGLCREWNL GSGKGSTVFE VYHAFCKASG 300
301 IDLPYKVTGR RAGDVLNLTA KPDRAKRELK WQTELQVEDS CKDLWKWTTE NPFGYQLRGV 360
361 EARFSAEDMR YDARFVTIGA GTRFQATFAN LGASIVDLKV NGQSVVLGYE NEEGYLNPDS 420
421 AYIGATIGRY ANRISKGKFS LCNKDYQLTV NNGVNANHSS IGSFHRKRFL GPIIQNPSKD 480
481 VFTAEYMLID NEKDTEFPGD LLVTIQYTVN VAQKSLEMVY KGKLTAGEAT PINLTNHSYF 540
541 NLNKPYGDTI EGTEIMVRSK KSVDVDKNMI PTGNIVDREI ATFNSTKPTV LGPKNPQFDC 600
601 CFVVDENAKP SQINTLNNEL TLIVKAFHPD SNITLEVLST EPTYQFYTGD FLSAGYEARQ 660
661 GFAIEPGRYI DAINQENWKD CVTLKNGETY GSKIVYRFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
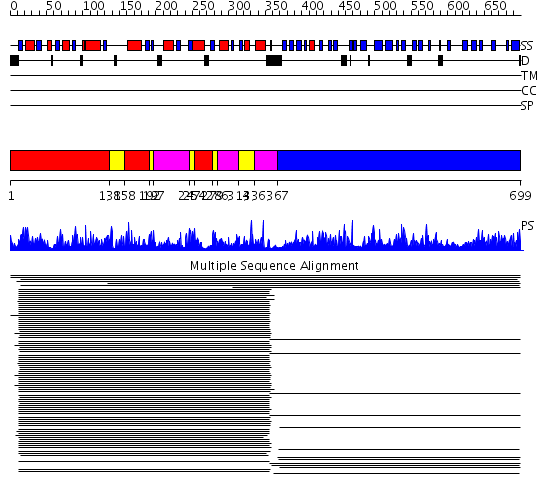
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] [158..191] [254..278] |
1160.0 | Uridine diphosphogalactose-4-epimerase (UDP-galactose 4-epimerase) | |
| 2 | View Details | [138..157] [192..196] [247..253] [279..285] [314..335] |
1160.0 | Uridine diphosphogalactose-4-epimerase (UDP-galactose 4-epimerase) | |
| 3 | View Details | [197..246] [286..313] [336..366] |
1160.0 | Uridine diphosphogalactose-4-epimerase (UDP-galactose 4-epimerase) | |
| 4 | View Details | [367..699] | 244.67837 | Galactose mutarotase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)