













| General Information: |
|
| Name(s) found: |
gi|157929160
[NCBI NR]
gi|157928632 [NCBI NR] |
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | synthetic construct |
| Length: | 623 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPKVTSELL RQLRQAMRNS EYVTEPIQAY IIPSGDAHQS EYIAPCDCRR AFVSGFDGSA 60
61 GTAIITEEHA AMWTDGRYFL QAAKQMDSNW TLMKMGLKDT PTQEDWLVSV LPEGSRVGVD 120
121 PLIIPTDYWK KMAKVLRSAG HHLIPVKENL VDKIWTDRPE RPCKPLLTLG LDYTGISWKD 180
181 KVADLRLKMA ERNVMWFVVT ALDEIAWLFN LRGSDVEHNP VFFSYAIIGL ETIMLFIDGD 240
241 RIDAPSVKEH LLLDLGLEAE YRIQVHPYKS ILSELKALCA DLSPREKVWV SDKASYAVSE 300
301 TIPKDHRCCM PYTPICIAKA VKNSAESEGM RRAHIKDAVA LCELFNWLEK EVPKGGVTEI 360
361 SAADKAEEFR RQQADFVDLS FPTISSTGPN GAIIHYAPVP ETNRTLSLDE VYLIDSGAQY 420
421 KDGTTDVTRT MHFGTPTAYE KECFTYVLKG HIAVSAAVFP TGTKGHLLDS FARSALWDSG 480
481 LDYLHGTGHG VGSFLNVHEG PCGISYKTFS DEPLEAGMIV TDEPGYYEDG AFGIRIENVV 540
541 LVVPVKTKYN FNNRGSLTFE PLTLVPIQTK MIDVDSLTDK ECDWLNNYHL TCRDVIGKEL 600
601 QKQGRQEALE WLIRETQPIS KQH |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
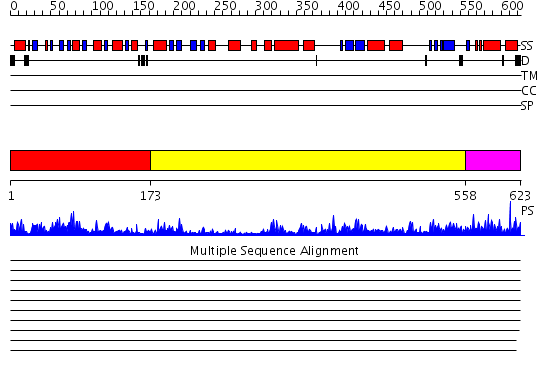
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 2.11 | Creatinase; Creatinase, catalytic (C-terminal) domain | |
| 2 | View Details | [173..557] | 75.522879 | Prolidase from Pyrococcus furiosus | |
| 3 | View Details | [558..623] | 40.30103 | Methionine aminopeptidase, insert domain; Methionine aminopeptidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)