













| General Information: |
|
| Name(s) found: |
gi|190406171
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 685 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFIALISGG KDSFYNIFHC LKNNHELIAL GNIYPKESEE QELDSFMFQT VGHDLIDYYS 60
61 KCIGVPLFRR SILRNTSNNV ELNYTATQDD EIEELFELLR TVKDKIPDLE AVSVGAILSS 120
121 YQRTRVENVC SRLGLVVLSY LWQRDQAELM GEMCLMSKDV NNVENDTNSG NKFDARIIKV 180
181 AAIGLNEKHL GMSLPMMQPV LQKLNQLYQV HICGEGGEFE TMVLDAPFFQ HGYLELIDIV 240
241 KCSDGEVHNA RLKVKFQPRN LSKSFLLNQL DQLPVPSIFG NNWQDLTQNL PKQQAKTGEQ 300
301 RFENHMSNAL PQTTINKTND KLYISNLQSR KSETVEKQSE DIFTELADIL HSNQIPRNHI 360
361 LSASLLIRDM SNFGKINKIY NEFLDLSKYG PLPPSRACVG SKCLPEDCHV QLSVVVDVKN 420
421 TGKEKINKNK GGLHVQGRSY WAPCNIGPYS QSTWLNDDAN QVSFISGQIG LVPQSMEILG 480
481 TPLTDQIVLA LQHFDTLCET IGAQEKLLMT CYISDESVLD SVIKTWAFYC SNMNHRSDLW 540
541 MDKSDDVEKC LVLVKISELP RGAVAEFGGV TCKRLIVDDN DSDKKEREEN DDVSTVFQKL 600
601 NLNIEGFHNT TVSAFGYNRN FITGFVDSRE ELELILEKTP KSAQITLYYN PKEIITFHHH 660
661 IGYYPVEKLF DYRGKEHRFG LHIRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
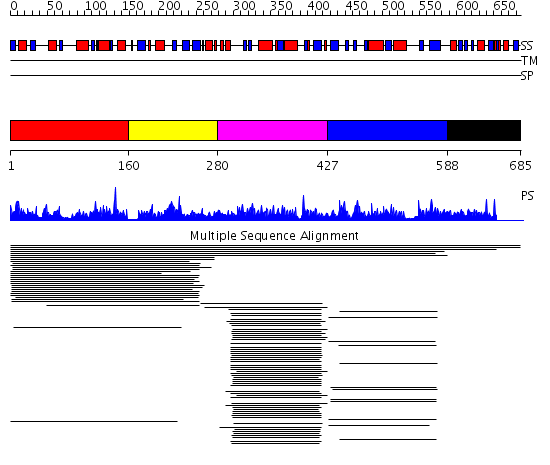
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 13.59 | Argininosuccinate synthetase, N-terminal domain; Argininosuccinate synthetase, C-terminal domain | |
| 2 | View Details | [160..279] | 11.275724 | ATP-binding region No confident structure predictions are available. | |
| 3 | View Details | [280..426] | 102.751666 | Purine regulatory protein YabJ | |
| 4 | View Details | [427..587] | 28.15299 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [588..685] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)