













| General Information: |
|
| Name(s) found: |
gi|170025189
[NCBI NR]
gi|169751723 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Yersinia pseudotuberculosis YPIII |
| Length: | 407 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVDINVPD LPESVADGSV ATWHKKPGDS VKRDEVLVEI ETDKVILEVP ASQDGILDAI 60
61 LEDEGATVTS RQVLGRIRPS DSSGKPTEEK SQSTESTPAQ RQTASLEEES NETLSPAIRR 120
121 LIAEHDLDAT AIKGSGVGGR ITREDVDSHL ASRKSASAVV ADAKAVAAAA PVLAGRSEKR 180
181 VPMSRLRKRV AERLLEAKNS TAMLTTFNEI NMQPIMDLRK QYGEAFEKRH GVRLGFMSFY 240
241 IKAVVEALKR YPEVNASIDG EDVVYHNYFD VSIAVSTPRG LVTPVLRDVD TLSMADIEKK 300
301 IKELAVKGRD GKLKVEELTG GNFTITNGGV FGSLMSTPII NPPQSAILGM HAIKDRPMAV 360
361 NGQVVILPMM YLALSYDHRL IDGRESVGYL VTVKEMLEDP ARLLLDV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
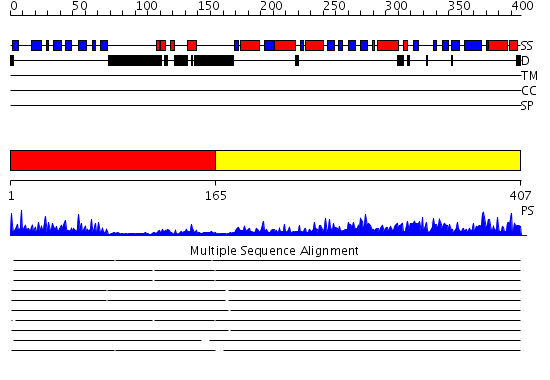
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 21.39794 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | |
| 2 | View Details | [165..407] | 89.69897 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)