













| General Information: |
|
| Name(s) found: |
gi|218553253
[NCBI NR]
gi|218360021 [NCBI NR] |
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli IAI1 |
| Length: | 405 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVDILVPD LPESVADATV ATWHKKPGDA VVRDEVLVEI ETDKVVLEVP ASADGILDAV 60
61 LEDEGTTVTS RQILGRLREG NSAGKETSAK SEEKASTPAQ RQQASLEEQN NDALSPAIRR 120
121 LLAEHNLDAS AIKGTGVGGR LTREDVEKHL AKAPAKESAP AAAAPAAQPA LAARSEKRVP 180
181 MTRLRKRVAE RLLEAKNSTA MLTTFNEVNM KPIMDLRKQY GEAFEKRHGI RLGFMSFYVK 240
241 AVVEALKRYP EVNASIDGDD VVYHNYFDVS MAVSTPRGLV TPVLRDVDTL GMADIEKKIK 300
301 ELAVKGRDGK LTVEDLTGGN FTITNGGVFG SLMSTPIINP PQSAILGMHA IKDRPMAVNG 360
361 QVEILPMMYL ALSYDHRLID GRESVGFLVT IKELLEDPTR LLLDV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
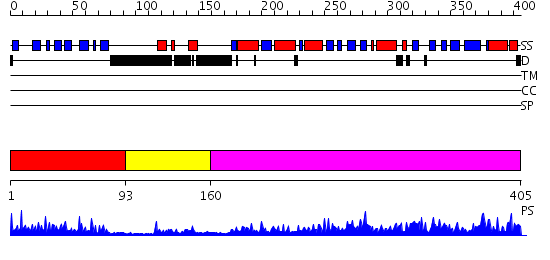
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 15.39794 | LIPOYL DOMAIN FROM THE DIHYDROLIPOYL SUCCINYLTRANSFERASE COMPONENT OF THE 2-OXOGLUTARATE DEHYDROGENASE MULTIENZYME COMPLEX OF ESCHERICHIA COLI, NMR, 25 STRUCTURES | |
| 2 | View Details | [93..159] | 4.0 | Solution structure of the e3_binding domain of dihydrolipoamide branched chaintransacylase | |
| 3 | View Details | [160..405] | 91.0 | Dihydrolipoamide succinyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)