













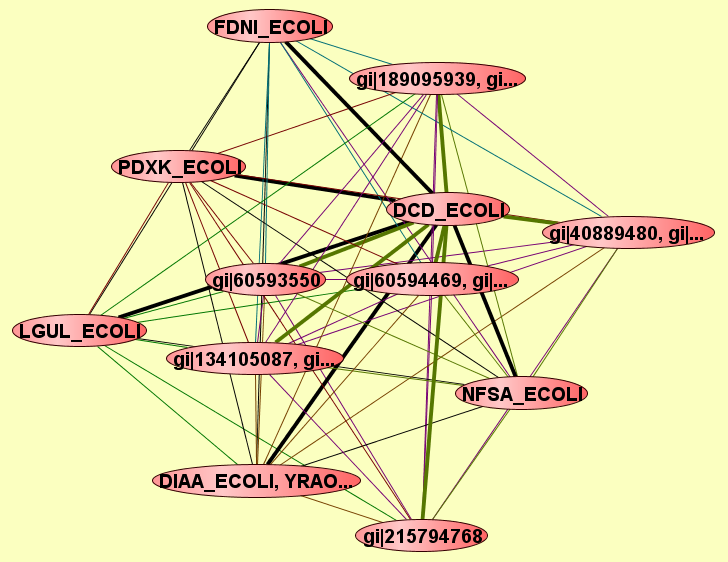
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1TUVA and 1XS1A with confidence: 0.922628612026 | DCD_ECOLI gi|60593550 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1F9ZA and 1XS1A with confidence: 0.974233049579 | DCD_ECOLI LGUL_ECOLI |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2YVAA and 1XS1A with confidence: 0.951063436312 | DCD_ECOLI YRAO_ECOLI, DIAA... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 3ERSX and 1XS1A with confidence: 0.982745963382 | DCD_ECOLI gi|215794768 |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2DDMA and 1XS1A with confidence: 0.952101056203 | DCD_ECOLI PDXK_ECOLI |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2PURA and 1XS1A with confidence: 0.998500885571 | DCD_ECOLI gi|189095939, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 2O9GA and 1XS1A with confidence: 0.995667446208 | DCD_ECOLI gi|134105085, gi... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1F5VA and 1XS1A with confidence: 0.959507810048 | DCD_ECOLI NFSA_ECOLI |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1XVIA and 1XS1A with confidence: 0.977652446278 | DCD_ECOLI gi|60594470, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1XS1A and 1QYNA with confidence: 0.947754372027 | DCD_ECOLI gi|40889477, gi|... |
| View GO Analysis | 2 proteins | Predicted to be cocomplexed based on PDB structures 1XS1A and 1KQFC with confidence: 0.985226444859 | DCD_ECOLI FDNI_ECOLI |