| |
Size |
Notes |
Members |
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1RWIA and 1SIXA with confidence: 0.932452976115 |
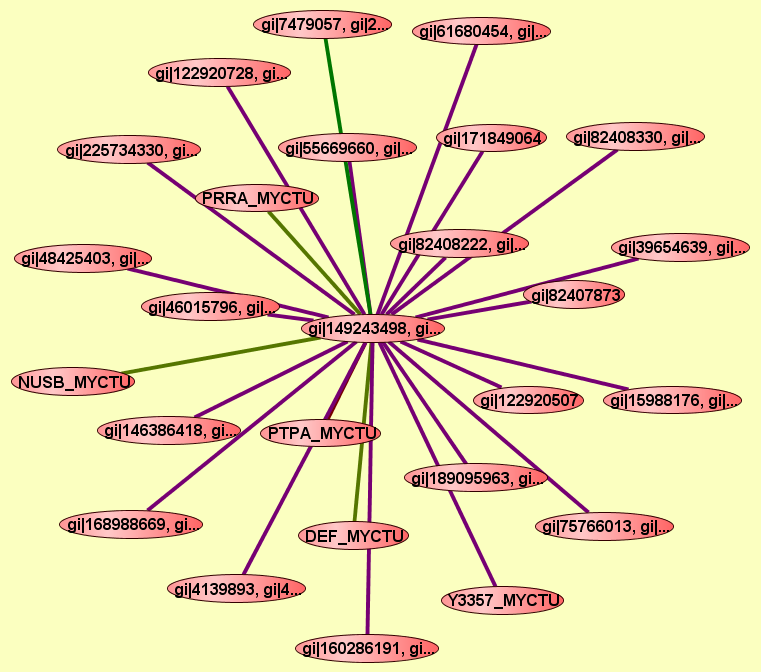
gi|149243498, gi...
gi|48425401, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1Q74A and 1SIXA with confidence: 0.905622900093 |
MSHB_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1L1EA and 1SIXA with confidence: 0.922434125659 |
CMAS3_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 2JEKA and 1SIXA with confidence: 0.903611167651 |
gi|149243498, gi...
gi|7459124, gi|1...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 3GMGA and 1SIXA with confidence: 0.930127609429 |
gi|149243498, gi...
gi|225734329, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1QPOA and 1SIXA with confidence: 0.932890752901 |
gi|149243498, gi...
gi|4139887, gi|4...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1EYVA and 1SIXA with confidence: 0.949377420712 |
NUSB_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 3D55A and 1SIXA with confidence: 0.966897522139 |
Y3357_MYCTU, REL...
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1RIIA with confidence: 0.940288979379 |
gi|149243498, gi...
gi|55669659, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2ASFA with confidence: 0.939555214549 |
gi|149243498, gi...
gi|82407873
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1SGVA with confidence: 0.935537047553 |
gi|149243498, gi...
gi|46015795, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1YS7A with confidence: 0.903535708144 |
PRRA_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2R5UA with confidence: 0.923937398097 |
gi|149243498, gi...
gi|189095965, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2IRUA with confidence: 0.900218307792 |
gi|122920722, gi...
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 3E3UA with confidence: 0.900956639593 |
DEF_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1U2PA with confidence: 0.934778556752 |
PTPA_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2VF2A with confidence: 0.908679954693 |
gi|149243498, gi...
gi|160286191, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2I1UA with confidence: 0.9273137214 |
gi|122920507
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2PMUA with confidence: 0.944704883078 |
gi|149243498, gi...
gi|168988666, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2C2XA with confidence: 0.94563317922 |
gi|149243498, gi...
gi|82408221, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 3BZQA with confidence: 0.936316812312 |
gi|149243498, gi...
gi|171849064
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1XVWA with confidence: 0.957056470528 |
gi|149243498, gi...
gi|61680454, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2A2JA with confidence: 0.917828172793 |
gi|149243498, gi...
gi|75766013, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 1I9GA with confidence: 0.946470259514 |
TRMI_MYCTU
gi|149243498, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1SIXA and 2CGHA with confidence: 0.924612647262 |
gi|146386419, gi...
gi|149243498, gi...
|