| |
Size |
Notes |
Members |
| View GO Analysis |
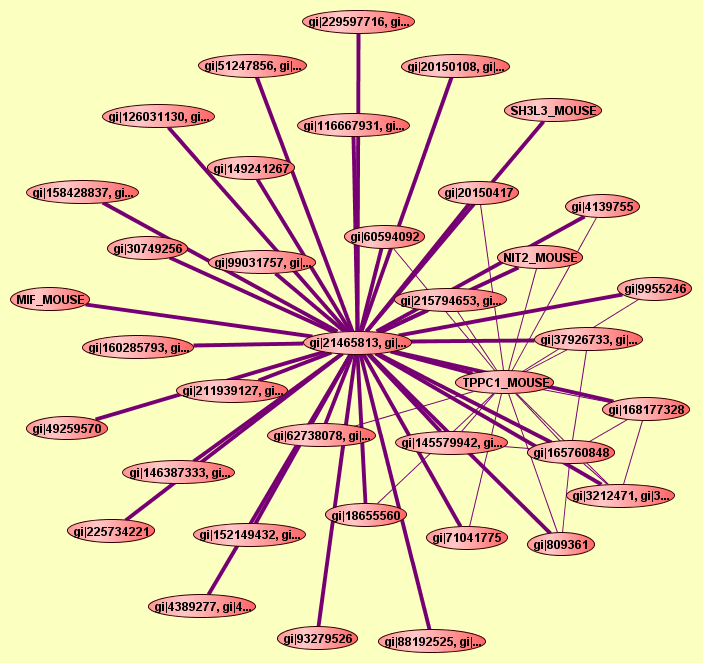
2 proteins |
Predicted to be cocomplexed based on PDB structures 1UYWH and 1KCVL with confidence: 0.957727710401 |
gi|21465813, gi|...
gi|62738080, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1VGKA and 1KCVL with confidence: 0.910890124426 |
gi|21465813, gi|...
gi|71041775
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1UUJA and 1KCVL with confidence: 0.901585600878 |
gi|21465813, gi|...
gi|51247855, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1GL2C and 1KCVL with confidence: 0.902931582613 |
gi|18655560
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1VKBA with confidence: 0.905746180311 |
gi|21465813, gi|...
gi|49259570
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2I13A with confidence: 0.922324591168 |
gi|116667931, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1I3ZA with confidence: 0.907719457291 |
gi|21465813, gi|...
gi|30749256
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2O27A with confidence: 0.962617328591 |
gi|145579947, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2J3TC with confidence: 0.991799754175 |
TPPC1_MOUSE
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1AY1H with confidence: 0.965875782519 |
gi|21465813, gi|...
gi|3212471, gi|3...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1BJ1H with confidence: 0.927381905445 |
gi|21465813, gi|...
gi|4389277, gi|4...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2GDGA with confidence: 0.923840175123 |
MIF_MOUSE
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2JCQA with confidence: 0.924512770348 |
gi|126031130, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1T1VA with confidence: 0.91269138597 |
SH3L3_MOUSE
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 3DURB with confidence: 0.917504035274 |
gi|21465813, gi|...
gi|215794651, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1JFQH with confidence: 0.902691790251 |
gi|20150417
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2Q2BA with confidence: 0.902854564686 |
gi|152149432, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1ZXKA with confidence: 0.903712773066 |
gi|21465813, gi|...
gi|99031757, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1XZZA with confidence: 0.912480592622 |
gi|21465813, gi|...
gi|60594092
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1I8KA with confidence: 0.926147436641 |
gi|20150111, gi|...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2C1PB with confidence: 0.946399215235 |
gi|21465813, gi|...
gi|88192524, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2E4FA with confidence: 0.914245935001 |
gi|149241267
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1DOWA with confidence: 0.940671699771 |
gi|21465813, gi|...
gi|9955246
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2IPUG with confidence: 0.918872222552 |
gi|158428837, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2W1VA with confidence: 0.916305967903 |
NIT2_MOUSE
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 3B5KA with confidence: 0.908518259197 |
gi|211939127, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 3C4BA with confidence: 0.973325449526 |
gi|168177328
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1EAPB with confidence: 0.943290595586 |
gi|21465813, gi|...
gi|809361
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2OYAA with confidence: 0.910532323403 |
gi|146387333, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2PSMA with confidence: 0.94988066365 |
gi|160285795, gi...
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2FIKA with confidence: 0.908924015068 |
gi|21465813, gi|...
gi|93279526
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1MJUL with confidence: 0.962537132053 |
gi|21465813, gi|...
gi|37926733, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 1WEJH with confidence: 0.943641409124 |
gi|21465813, gi|...
gi|4139755
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 2EH7L with confidence: 0.980160457185 |
gi|165760848
gi|21465813, gi|...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 3D14A with confidence: 0.906427005928 |
gi|21465813, gi|...
gi|237640459, gi...
|
| View GO Analysis |
2 proteins |
Predicted to be cocomplexed based on PDB structures 1KCVL and 3GBWA with confidence: 0.903494561319 |
gi|21465813, gi|...
gi|225734221
|