













| General Information: |
|
| Name(s) found: |
CG11423-PA /
FBpp0086012
[FlyBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial respiratory chain complex I
[ISS]
|
| Biological Process: |
oxidation reduction
[IEA]
|
| Molecular Function: |
FMN binding
[IEA]
NAD or NADH binding [IEA] NADH dehydrogenase (ubiquinone) activity [ISS] 4 iron, 4 sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISFLLKRRA CDAYKISLGL TRAYRALTEV KSLQNALRPG NKTGNIKLVN IYRTSSIQNV 60
61 KKNQLSDAES VIPLLLRPAK PVLIMREGSQ TDCNDDLLSS EQRAKKRESI KKSEVDCPKF 120
121 PAEKLPPTTK FPINENAKKV LDPADQRELE ANLLKRLKDM AKRKKSRLDE KVDELRDNFI 180
181 GWNYAALQGR TIVTVDAKES TPPKGSGPAT PAGSPPPKNV SAPPTGGSPP AKSGAPPPPK 240
241 GPPPGTPPPQ TKTTFGPLAD ADRIFTNLYG RHDWRLKAAM KRGDWYKTKE IIAKGDKWIV 300
301 NEIKTSGLRG RGGAGFPSGL KWSFMHKPPD GRPKFLVVNA DEGEPGTCKD REIMRHDPHK 360
361 LVEGCLIAGR AMGANTGFIY IRGEFYNEAC NLQYAIIEAY KAGYLGKNAC GSGFDFDLYV 420
421 QRGAGAYICG EETSLIESLE GKAGKPRNKP PFPADIGVFG CPSTVTNVET VAVSPTICRR 480
481 GGNWFASFGR TRNSGTKLFN ISGHVCNPCT VEEEMSMPTR ELIERHAGGV IGGWDNLLAI 540
541 IPGGSSTPCI TKEHASSAIH DYDGLMAVRS SMGTGALIVM NKDTDIIKAI ARLSAFYKHE 600
601 SCGQCTPCRE GLHWVNMIMH RFVTGQAQIA EIDMLFELTK QIEGHTICAL ADGAAWPPQG 660
661 LIRNFRPVIE ERIKKRAAED QAKAEALEAE RFPCQTVTPC PE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
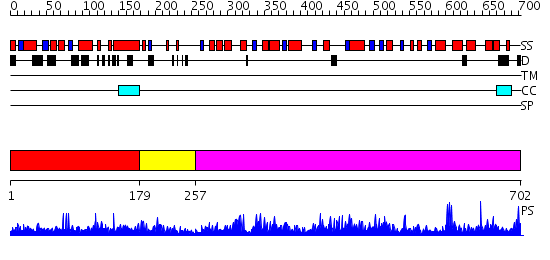
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [179..256] | 1.175987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [257..702] | 157.0 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)