













| General Information: |
|
| Name(s) found: |
FBpp0311230
[FlyBase]
ca-PA / FBpp0084850 [FlyBase] |
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1961 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
eye pigment precursor transport
[TAS]
ommochrome biosynthetic process [IMP] eye pigment granule organization [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGQAELKEL GMFELRPLIT NISCKLSAVT AGLDEKRQFL CLVSEENEIL LRYISAASGQ 60
61 EVVKMISWFR NRVIHDVCFD PAGVWLLVLC FDNTLHIVPA LALVDKAHGL SACSLYSTTQ 120
121 VTSYIIPFVG PHECPNSKKC PNHSTGLEES TAEPMDQLAE NLEQARLQEG EEEEPLGHDL 180
181 PEHMLVRNSG TEDAVQPLPQ QQQTIDTTGN SQVMAASFMA SKTCPYPLSV VWWKTNSGQN 240
241 RAIIGYSDGS ICFVGLSPNC PMIASTAIEN GSVMRLVICK DKAYDGVMLL ITSSLKQQYK 300
301 LLLEQKAIKY VYPGDSSPTT REGAWQIVMS VQDKHQSAAT GARQSNSSDP ASDSSQLEED 360
361 VFAPPSSDDV VCSGPGAANS DPSTVSASAE AATADVPPPP APPSYSEAVA RQSGDQGGVR 420
421 FQQQQLPAGS MDGILPATRA RLASLKSLGS KKLQAIKMRL SDQRQKFDEF MPDSTMGSFA 480
481 IMDSPSVTPE PLGTTSGSFY NIQNLRSTFL LSALHSNSHS LTVHSIDISL KPLFVYKIPK 540
541 HTQEILISSN ILYGIHYVDL HCRSDSDHFT AASLLDDPLV ANTEKENRDA DIEKVDQSDV 600
601 SAQKSDSTSA TTTATSSLDQ SDMPSTAINA VSVISSAMAS LHTSDEDRFN NLAQLGLFRF 660
661 ENETVLHLYQ MAQYRNPSAE PETLTKEEKA KAFELKDIHT PMDYIQFYET ADVSSEFSLR 720
721 QMEIMQSEFP KINYDPSYVI TNKNVYTIQL KKEPFELFLD AALQNEFNQC RDFCNTFDLS 780
781 FEQCIEFAGD YLLRRKKITQ ALLTYNKARV KPIRTALKLA MYGHTYALMQ LSAMALKSTY 840
841 LLRSRYLGHP LLKGILGDIT YRHSEDVRVI LPEKHEEEDV NSGVFCSDYH YGVDESISAL 900
901 QMSPSSQFHL ANLLLITLAE KAVNDKNYIP LWNFLVTNSK YHTNMTSIVL CQSGLFSSAI 960
961 ILAKIRGVCL DTFNALISVV AQEFGWYNEL NVCLYNLSEN IFLETITYLP HVSMDYFAFI1020
1021 QEKLHHIRPC VLQRLSKQLN PFSPALRPIV SHNSDCAMSN DNNPQLFEFC KSLIETYLAV1080
1081 LIHMESQRVK YDSIVNALSN FRINYEDNQF AIESSRFKPI SAGCAYCACV VDGTAYFWGS1140
1141 SGIPSYYSAQ KSTEPPEPAH AVKSLDLLSQ LNLQVHAIKC GRQHTLILTN NGLYSLGNNN1200
1201 LCQLGIGRHM QMALQPMLVT ALDGMNITML EAGQYHNAAV ADGKLYMWGW GIYGQLGQGS1260
1261 CENIATPQLV SFFKFKKILQ ISLGHAHTLV LCGAPNSHME ANHCGNELFV FGSNHFGQLG1320
1321 TGNSDHGDTK TNLPVRLEVG DCSLRLIHTK FFTNLAVDEQ DRLFTWGSSP QALRLANQIK1380
1381 RRANAKQKME ENQQRELLSK QLEATLLPAP IPSTSTLVEP TTSYAAVVAG STPKSSVSDT1440
1441 KDCQDVQNEA APSDATETIA PASAKAAPPP VPVASPPVLV PEPDPTEHLT PHLVDTSEVA1500
1501 GRILQISSGL FHFCLISSSC TLYTWGKNLE HQLGTEDKDR RAVLKPSPID SIERPMYVDC1560
1561 GSDFTLVMTT DHVVRAFGGN SNGQCGRDLG PSVERMRQRV VCLPATKRLV RFESQCVEVP1620
1621 VEINLPRPRI RLDQDPVRYL KVMPPYQPHF LQPANISFDQ HCSVGTPNYK SSTEHAATGQ1680
1681 DSDDMLSSLE NLSQNDASFS YNAPVLGDAH SSLDMDYTPE EQSYVPLPEQ AVGSGMPANA1740
1741 EQLLGRDDDD GSTYTQSTEG MGESSLQHLR HYIHYCLYAF HGLYNPDKIG ECARPHRNEY1800
1801 RIRICMLNFR FVEAFRLCLQ SCGSAQRTLK LFEYFSKDTG YVPLRRTDLK HLIYHLCLHF1860
1861 LERKFDMLEC ERFFMGDLDY YLLELAYVFY FNNNNSTLEQ NLNQKFKQLI AAADSNQAAT1920
1921 SAGQELEQLQ DTDVIFDSFT VKFKTILCQR LLSYSDCDAA N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
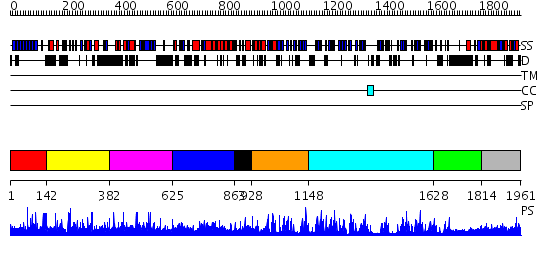
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..381] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [382..624] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [625..862] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [863..927] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [928..1147] | 33.09691 | beta-lactamase inhibitor protein-II, BLIP-II | |
| 7 | View Details | [1148..1627] | 84.09691 | Regulator of chromosome condensation RCC1 | |
| 8 | View Details | [1628..1813] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1814..1961] | 1.040995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 |
|
|||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)