













| General Information: |
|
| Name(s) found: |
l(3)82Fd-PG /
FBpp0089229
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1033 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell wall macromolecule catabolic process
[IEA]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNVILTSQY YWSRRASQDV SSLSRSVDNL AIPVRRSKSR SVDHGLAAPF DLDSLRSKVE 60
61 QRFESVDKLS RQKSSLPTIP TISYTVGNRD TLTSVAARFD TTPSELTHLN RLNSSFIYPG 120
121 QQLLVPDKSA KDDASSSSTH DADGASLSGK SSPVERKLSG DESREKDILE GLRPGSPKPG 180
181 HIERVGGSSQ QQQAEEEANK SDDPVITQRF LKINVRHITD GQGVVGGVLL VTPNAVMFDP 240
241 NVSDPLVIEH GPESYGVIAP MELVVNAAIF HDIAHMRVAG GAGQGLNSAS GDAEKPEIYY 300
301 PKPVLVEEDS KELPEQQSLL GEDGNKRLDA EIGSLEIADD QESLCSSTGR DGDAFPKAFD 360
361 RERVEDSSME AKDSAKTDAL DDEDKKAGLG LSSTRSTLEE RRKSLLDHHW AIPSKDRYCL 420
421 SNSRRSSEDE GDNESNVTVD SGARVQDPHS ANSSVSGVPL SQPPVAAAAA AAAVAPASAV 480
481 VPLPPSGIDL EHLEQLSKQS CYDSGIDIRE PVPNVQPIPK KTVYSDADIV LSSDWVPPKT 540
541 IVPTHFSESP PRSTILGQSL DAGTGGAGGV RKKTSSVSFS VDDEAAQQAQ AQSAALAVTD 600
601 KQADKKNKML KRLSYPLTWV EGLTGEGGAV PPGGVGSGSL NKSGDTDSAP NTGDSNQSVF 660
661 SKVFSRRSSI GTFIRPHSSE GTSSSTKLKE AKQAPKLDYR SMVSMDDKPE LFISVDKLIP 720
721 RPARACLDPP LYLRLRMGKP IGKAIPLPTS VMSYGKNKLR AEYWFSVPKN RVDELYRFIN 780
781 TWVKHLYGEL DEEQIKARGF ELIQEDTEWT KSGTTKAGMG GGSQDGEEIS DLTRESWEVL 840
841 SMSTDEYRKT SLFATGSFDL DFPIPDLIGK TEILTEEHRE KLCSHLPARA EGYSWSLIFS 900
901 TSQHGFALNS LYRKMARLES PVLIVIEDTE HNVFGALTSC SLHVSDHFYG TGESLLYKFN 960
961 PSFKVFHWTG ENMYFIKGNM ESLSIGAGDG RFGLWLDGDL NQGRSQQCST YGNEPLAPQE1020
1021 DFVIKTLECW AFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
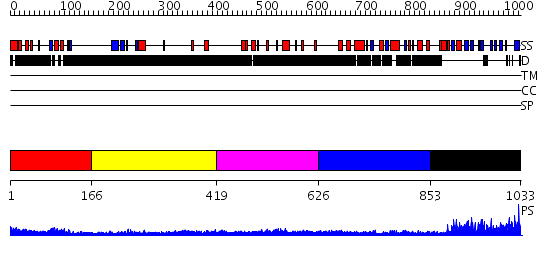
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 3.5 | No description for 2djpA was found. | |
| 2 | View Details | [166..418] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [419..625] | 1.25799 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [626..852] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [853..1033] | 10.131983 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)