













| General Information: |
|
| Name(s) found: |
Pi4KIIalpha-PC /
FBpp0089179
[FlyBase]
Pi4KIIalpha-PA / FBpp0089178 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
phosphoinositide phosphorylation
[IDA]
|
| Molecular Function: |
1-phosphatidylinositol 4-kinase activity
[IDA][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGARDQTDP PLLQLEDEVD LIGDFAKDQP PPRNQLSPLL NGFESTSNQL TTKVEINTVG 60
61 MGKRKTRRRG KKAREPMLLS VDDEDDEEEA QEERNHTNGL PPGEESAGGD AVIQKPQLDN 120
121 LVMHYDDQID DEDGEEAVLL KRSSNPRNQR LKGSAEDSID SSSQQPPIIC HIAPAKVASS 180
181 ALTSPLKTTS HTALARHPHD EADDICLVPE IDFEGMSTDA GSDNRESQPL LGGGNASHGS 240
241 RISELLGGLG RGHDHVEFVS NTFGDDPLFS EIVAKAEGAI ENGVLPERIY QGSSGSYFVK 300
301 DASHQCLAVF KPKDEEPYGR LNPKWTKWMH KLCCPCCFGR ACLIPNQGYL SESGASLVDR 360
361 KLNLNVVPKT RVVRLVAESF NYARIDRQKA KLKKRIKEHY PSAHFNRMSL PLKTGSFQLF 420
421 VEGYKDADYW LRRFESEPLP PSVAKSFQLQ FERLVVLDYI IRNTDRGNDN WLIKYVAPKI 480
481 TTQVGGTGGR TMHGVVTPKL PNALGASKIV DLNEPPQASP KPTTGGRPSN HLAGGAATSA 540
541 QQDSSSLASS EDEPSSRSPP TSEPDWNVVN SAFIRIAAID NGLAFPFKHP DSWRAYPYHW 600
601 AWLPQAKIPF SEEIKEQVLP QLSDMNFVEE ICTELLYLFQ QDRGFDKRLF ERQMSVMRGQ 660
661 ILNLTQALRD GKSPVQLVQM PAVIVERSSG SRFFSFTQRF QNKSPFFSWC |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
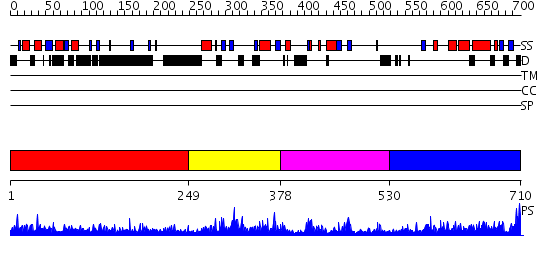
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [249..377] | 3.055967 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [378..529] | 3.058994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [530..710] | 3.057929 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)