













| General Information: |
|
| Name(s) found: |
FBpp0301185
[FlyBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 777 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
purine base metabolic process
[IEA]
purine ribonucleoside monophosphate biosynthetic process [IEA] |
| Molecular Function: |
AMP deaminase activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNFNRMDTAH RLRVDVRLYG GSMGNQLNDK GTASSTPTIS SDDLSDLADS PVDEKAEADV 60
61 EVTNEISAPY EVPQFPIEQI EKKLQIQRHL NEKQATGPRP VATAAVLATN RESSSSTEGR 120
121 ESAVTMERND ADINFQRVSI SGEDTSGVPL EDLERASTLL IEALRLRSHY MAMSDQSFPS 180
181 TTARFLKTVK LKDRINNLPV KEVSESADVH LRHSPMKITN PWNVEFPNDE DFKIKPLNGV 240
241 FHIYENDDES SEIKYEYPDM SQFVNDMQVM CNMIADGPLK SFCYRRLCYL SSKYQMHVLL 300
301 NELRELAAQK AVPHRDFYNT RKVDTHIHAA SCMNQKHLLR FIKKTLKNNA NEVVTVTNGQ 360
361 QMTLAQVFQS MNLTTYDLTV DMLDVHADRN TFHRFDKFNS KYNPIGESRL REVFLKTDNY 420
421 LNGKYFAQII KEVAFDLEES KYQNAELRLS IYGKSPDEWY KLAKWAIDND VYSSNIRWLI 480
481 QIPRLFDIFK SNKMMKSFQE ILNNIFLPLF EATARPSKHP ELHRFLQYVI GFDSVDDESK 540
541 PENPLFDNDV PRPEEWTYEE NPPYAYYIYY MYANMTVLNK FRQSRNMNTF VLRPHCGEAG 600
601 PVQHLVCGFL MAENISHGLL LRKVPVLQYL YYLTQIGIAM SPLSNNSLFL NYHRNPLPEY 660
661 LARGLIISLS TDDPLQFHFT KEPLMEEYSI AAQVWKLSSC DMCELARNSV MMSGFPHAIK 720
721 QQWLGPIYYE DGIMGNDITR TNVPEIRVAY RYETLLDELS NIFKVNQTCS IADMNAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
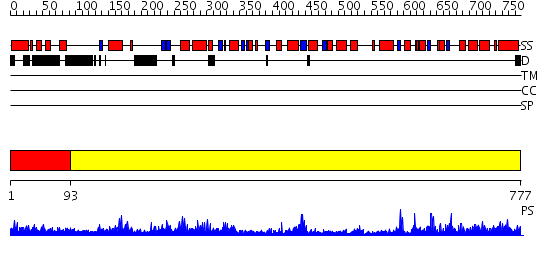
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [93..777] | 174.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)