













| General Information: |
|
| Name(s) found: |
Crag-PB /
FBpp0089320
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1644 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell cortex
[IDA]
recycling endosome [IDA] |
| Biological Process: |
basement membrane assembly
[IMP]
asymmetric protein localization [IMP] |
| Molecular Function: |
calmodulin binding
[IDA]
guanyl-nucleotide exchange factor activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEEKRIADYF VVAGMPENPQ LLQENSFNDS GRLRAATTIE PITDIGVYFP LLGEEVPEGY 60
61 EILSHTPTGL QANLNHGSVR TTDCYIYFRR GKDRPPLVDI GVLYDGHERI MSDAEIVAET 120
121 PGGRVANVNN SSAKTFLTYR RARADMPCNE LVVTELCVIV QSKGERAPHA FCLIYKTLNK 180
181 GYVGSDVYLC YKKSMYRPKH ISYKPEILLR YPTFDHTDFP LNLCPSVPLF CLPMGASLEA 240
241 WPHVNGTEKR KPISPVFSTF VLTVSDGTYK VYGSALTFYE DYDESKLSAE QKELLGWDEE 300
301 FGAQHSLHMI KAICLLSHHP FGDTFDKWLK YLHRMVLYDV NIPIPVERYI TQLLDEVPFP 360
361 APSIHLQLSS ESNDRILLTQ PEDSPLPRSG AGFHILLQNL GTDNCLHVLL LALTEQKILI 420
421 HSLRPATLTA VAEAIVSLLF PFKWQCPYIP LCPLGLAEVL HAPLPYLIGV DSRFFDLYEP 480
481 PTDVTCIDLD TNNISLCESQ RHLSPKLLPK RAARLLRQTL TELENAKPIS YDSTNSLDRD 540
541 IRKRKRELVL EQRIQEAFLL FMASILRGYR DFLVPISKAP SVGATDPSAL FQLKAFLRSR 600
601 DKSHQKFFEL MMKTQMFIRF IEERSFVSDG DHGLSFFDEC AEKVGNYDET PAQLHLVDWD 660
661 TGQNSERTKY IFPPDSVTPA GSQPGGGGGG LAYNYENFTL QPELLQSTKK TALSKFLQLQ 720
721 LNASLSPGSP IARRTKHEIK LSQKMASRCQ QHPEAWSKYL LATCYSLYFL ILPSMVLDPR 780
781 HAGKEPEILR AAYDVLVRAS RLKITCDEFC YRIMMQLCGI HNLPVLAVRL HYLMKRSGVQ 840
841 ANALTYGFYN RCVLESQWPQ DSTTISQIRW NRIKNVVLGA AQFRKAGKQR AASKVNKSLS 900
901 ASQDQNLSTL ETVDGQSRTS LASSSGDGGG HGLLDFAAFD RLRNKLGSIV RQTVTGGNNE 960
961 TMGDAVNNTG LLIPGELSAP NTPTYGGDTE ILAKALQQQQ PRKQIMSIGG GDDDDEDEDE1020
1021 DEYTAGSPST PQKQLEAGAD DLEYAGGGDY EADEEDEDEV DEHVAAQRAR QRVQSPTKIS1080
1081 PRTPVTQNDP LGALNEEESA ASATPTQETQ QEQQHSQSQI DSSIYSDKPI LFRGQRSATF1140
1141 DESTQIGKSM HRSETMPVAS SGVTNSLANI GSSLKFTFGP SLTGKKSNEL IQGSLSSIKS1200
1201 AANSLTKKFD EIKGVISANS TPTKTNNGHH PHGLHHGHHH PHHHHHHHSQ HGNAEQEEHD1260
1261 AAVHEEGKLR RVSSDLDPWG RLSESRKSSY NNLVPLGENS STGALHMHAF PAVPDNLYSL1320
1321 TSENAADRDC DVLIQLTTCS QCHNCSVLVY DEEIMSGWTA EDSNLNTTCH ACNKLTVPFL1380
1381 SVQIERQVEE SEQSDPLQDG KEQIANGNSH KTSLTVPYLN PLVLRKELEN ILTQEGDIAL1440
1441 IKPEFVDEHP IIYWNLLWLM ERIESKTHLP ELCLPVPSDK EHIDPLSKVK TVHIQCLWDN1500
1501 LSLHTEASGP PMYLLYRETQ PTSPLIKALL TDQAQLNKNV IQQIISAIRC NDFATPLKRL1560
1561 ANERHKLKSN GVERSHSFYR DILFLALTAI GRSNVDLATF HREYAAVFDK LTERECNMYY1620
1621 RNQDLPPSAS TIFCRAYFRP LLLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
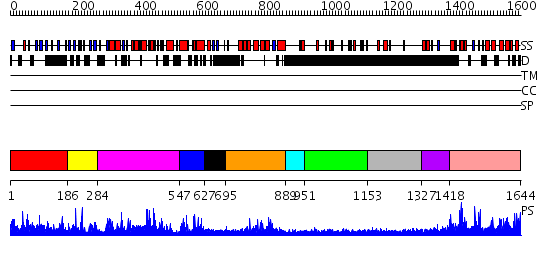
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..185] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [186..283] | 3.107905 | No description for PF03456.9 was found. No confident structure predictions are available. | |
| 3 | View Details | [284..546] | 66.886057 | No description for PF02141.12 was found. No confident structure predictions are available. | |
| 4 | View Details | [547..626] | 34.200659 | No description for PF03455.10 was found. No confident structure predictions are available. | |
| 5 | View Details | [627..694] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [695..888] | 20.105987 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [889..950] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [951..1152] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1153..1326] | 4.254988 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1327..1417] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1418..1644] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)