













| General Information: |
|
| Name(s) found: |
LIAS
[HGNC (HUGO)]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 372 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLRCGDAAR TLGPRVFGRY FCSPVRPLSS LPDKKKELLQ NGPDLQDFVS GDLADRSTWD 60
61 EYKGNLKRQK GERLRLPPWL KTEIPMGKNY NKLKNTLRNL NLHTVCEEAR CPNIGECWGG 120
121 GEYATATATI MLMGDTCTRG CRFCSVKTAR NPPPLDASEP YNTAKAIAEW GLDYVVLTSV 180
181 DRDDMPDGGA EHIAKTVSYL KERNPKILVE CLTPDFRGDL KAIEKVALSG LDVYAHNVET 240
241 VPELQSKVRD PRANFDQSLR VLKHAKKVQP DVISKTSIML GLGENDEQVY ATMKALREAD 300
301 VDCLTLGQYM QPTRRHLKVE EYITPEKFKY WEKVGNELGF HYTASGPLVR SSYKAGEFFL 360
361 KNLVAKRKTK DL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
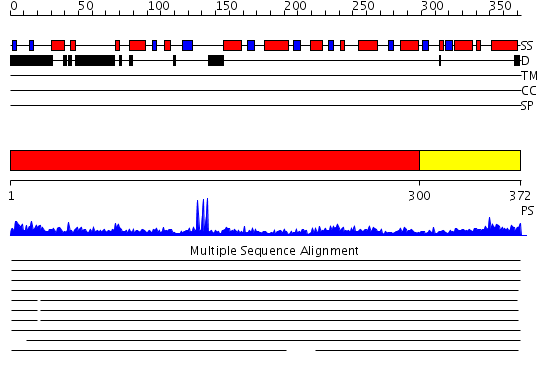
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 14.30103 | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | |
| 2 | View Details | [300..372] | 42.522879 | The Crystal Structure of Biotin Synthase, an S-Adenosylmethionine-Dependent Radical Enzyme |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)