













| General Information: |
|
| Name(s) found: |
YQIG_ECOLI
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Escherichia coli |
| Length: | 821 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGNIGANPV IIIGCASAYA VEFNKDLIEA EDRENVNLSQ FETDGQLPVG KYSLSTLINN 60
61 KRTPIHLDLQ WVLIDNQTAV CVTPEQLTLL GFTDEFIEKT QQNLIDGCYP IEKEKQITTY 120
121 LDKGKMQLSI SAPQAWLKYK DANWTPPELW NHGIAGAFLD YNLYASHYAP HQGDNSQNIS 180
181 SYGQAGVNLG AWRLRTDYQY DQSFNNGKSQ ATNLDFPRIY LFRPIPAMNA KLTIGQYDTE 240
241 SSIFDSFHFS GISLKSDENM LPPDLRGYAP QITGVAQTNA KVTVSQNNRI IYQENVPPGP 300
301 FAITNLFNTL QGQLDVKVEE EDGRVTQWQV ASNSIPYLTR KGQIRYTTAM GKPTSVGGDS 360
361 LQQPFFWTGE FSWGWLNNVS LYGGSVLTNR DYQSLAAGVG FNLNSLGSLS FDVTRSDAQL 420
421 HNQDKETGYS YRANYSKRFE STGSQLTFAG YRFSDKNFVT MNEYINDTNH YTNYQNEKES 480
481 YIVTFNQYLE SLRLNTYVSL ARNTYWDASS NVNYSLSLSR DFDIGPLKNV STSLTFSRIN 540
541 WEEDNQDQLY LNISIPWGTS RTLSYGMQRN QDNEISHTAS WYDSSDRNNS WSVSASGDND 600
601 EFKDMKASLR ASYQHNTENG RLYLSGTSQR DSYYSLNASW NGSFTATRHG AAFHDYSGSA 660
661 DSRFMIDADG TEDIPLNNKR AVTNRYGIGV IPSVSSYITT SLSVDTRNLP ENVDIENSVI 720
721 TTTLTEGAIG YAKLDTRKGY QIIGVIRLAD GSHPPLGISV KDETSHKELG LVADGGFVYL 780
781 NGIQDDNKLA LRWGDKSCFI QPPNSSNLTT GTAILPCISQ N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
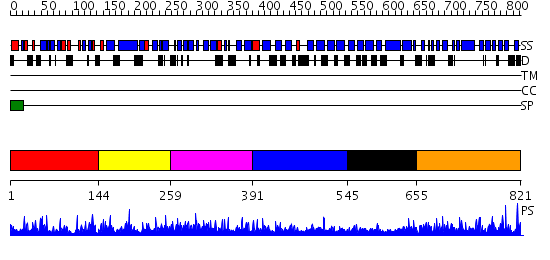
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 28.522879 | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | |
| 2 | View Details | [144..258] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [259..390] | 2.293979 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [391..544] | 1.298991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [545..654] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [655..821] | 3.295973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.63 |
Source: Reynolds et al. (2008)