













| General Information: |
|
| Name(s) found: |
SMC_MYCLE
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Mycobacterium leprae |
| Length: | 1203 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYLKSLTLKG FKSFASPTTL RFEPGITAVV GPNGSGKSNV VDALAWVMGE QGAKTLRGSK 60
61 MEDVIFAGTL SRAPLGRAEV TLIIDNSDNV LPIEYSEVSI TRRMFRDGAS EYEINGSSCR 120
121 LMDVQELLSD SGIGREMHVI VGQGKLDQIL QSRPEDRRTF IEEAAGILKY RRRKEKALRK 180
181 LDAMSANLAR LTDLTTELRR QLKPLSRQAE VARRAATIQA DLRDARLRLA ADDLVSRQGQ 240
241 RDAIVEAETM MRRDHDEAAA RLAVASEELA AHEAALTELS GRAESVQQIW FGLSTLVERV 300
301 SATIRIAGER AYHLDVDPAT PSDTDPDVLE AEAQQMEVAE QQLLAELAVA RTQLEAARAE 360
361 LADRERHAVE ADKAHLEAVR AEADRREGLA LLAGQVETMR ARIESIDDSV ARLSERIEEV 420
421 TARTQQILAE FETVQGRVGE LDQSEVHLDE QHERAVAALR FANERVAELQ SAERDAERQV 480
481 VSLRARIDAL TMGLERKDGA AWLARNYSDT GLLGSIAKLV KVRPGYEAAL AAVLGPAADA 540
541 LAVDSLGAAR SALTALKEAD AGRATLVLAD WLADAGPACV TGLPDGAQRA LDLIEAPPWL 600
601 QGALIAMLYG VVVVNYLAEA LGVVDICPQL RVVTVDGDLV GAGWVSGGSG RRLSTLEVTS 660
661 EIDKAGAELA AAEAHMAQLN AALSGALSEQ VAHSDATEQA LVALNESDTA ILSMYDQLGR 720
721 LGQEVRAAEA EWESLLAQRE ELEARRVSIL EEVVELETRL HNVEQIQHVH ALDENSAAAR 780
781 QLIVAAAEEA RGVEVEALLA VRTAEERVNA VCGRANSLRR AAAAEREVRL RDQQAHAARI 840
841 RAAAVAAAVT DCGQLLASRL TQAVDLAARH RDALATERQQ RSVAIAAVRY EVNTLRVRLA 900
901 TLTDSLHRDE VANVQAALRI EQLEQLVLEQ FGIAPVDLIA EYGPQVALLP TELEMAEFQQ 960
961 ARERGEQVTS PAPMPYDRAT QERRAKRAER ELAELGRVNP LALEEFAALE ERYNFLSTQL1020
1021 EDVKGARKDL LDVVAEVDAR ILQVFSDAFV DVEREFRGVF TSLFPGGEGR LRLTDPDDML1080
1081 TTGIEVEARP SGKKVSRLSL LSGGEKSLIA VAMLVAIFKA RPSPFYIMDE VEAALDDVNL1140
1141 CRLIGVFEQL RGQSQLIIIT HQKPTMEVAD TLYGVTMQGD GITAVISQRM RGQQVESLVT1200
1201 SSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
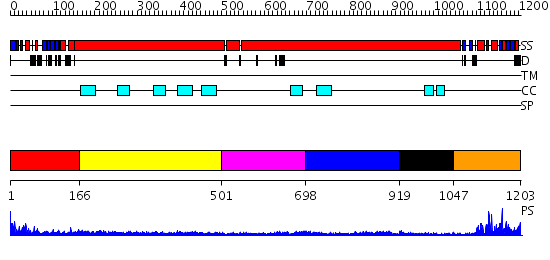
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..165] | 41.221849 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [166..500] | 2.99 | Tropomyosin | |
| 3 | View Details | [501..697] | 4.86 | Smc hinge domain | |
| 4 | View Details | [698..918] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [919..1046] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1047..1203] | 17.221849 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)