













| General Information: |
|
| Name(s) found: |
gi|151943715
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 917 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNVDERSRIG GREKDAGPGK GILKQNQSSQ MTSSFLENPG VRIPTRIITK KEVLDGSNTT 60
61 SRINTSNLQS MVKRRVSFAP DVTLHSFTFV PEQNNEIKEP RRRKTSTNSP TKISSQEEPL 120
121 VTSTQIDDAR TEEKTAAEED PDTSGMELTE PIVATPDSNK ASQHDPTSME MTEVFPRSIR 180
181 QKNPDVEGES IESSQQIDDV EAVREETMEL TAIHNVHDYD SISKDTVEGE PIDLTEYESK 240
241 PYVPNSVSRS TGKSSDYSVE RSNDKSDLSK SENKTNSSQP MEITDIFHAD PQNPMSLHSD 300
301 NNINNDGNEM ELTQIQTNFD RDNHHIDESP SEKHAFSSNK RRKLDTVSDY AASVTTPVKE 360
361 AKDTSGEDND GDLEMMEKMS PITFSDVDNK IGTRSNDVFT IEPGTEDTGM QTATDDEEDG 420
421 ENVDDNGNKI VEKTRLPEID KEGQSGIALP TQDYTLREFI NEVGVGFLDT KLIDDLDKKV 480
481 NFPLNSFNFV ENQRIDNVFS AFYIDIPILE VEAFRCKELW RSINESKDKF KDFEAQIDKS 540
541 HPPLLLQEYF SSDEKMKQLM RDQLQLVKGY SKLEAAMEWY EWRKKQLNGL ELILAENLNT 600
601 LKREYEKLNE EVEKVNSIRG KIRKLNEAIK EEIRSLKNLP SDSYKPTLMN RIKIEAFKQE 660
661 LMEHSISLSS SNDFTQEMRS LKLAIAKKSN DILTLRSEVA SIDKKIEKRK LFTRFDLPKL 720
721 RDTLKILESL TGVRFLKFSK ATLSIAFLQL DDLRVDINLA NFKNNPLSSM KVMNDSNNDD 780
781 MSYHLFTMLL KNVEAEHQDS MLSNLFFAMK KWRPLLKYIK LLKLLFPVKI TQTEEEEALL 840
841 QFKDYDRRNK TAFFYVISLV SFAQGVFSEN GQIPMKVHIS TQQDYSPSRE VLSDRITHKI 900
901 SGVLPSFTKS RIHLEFT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
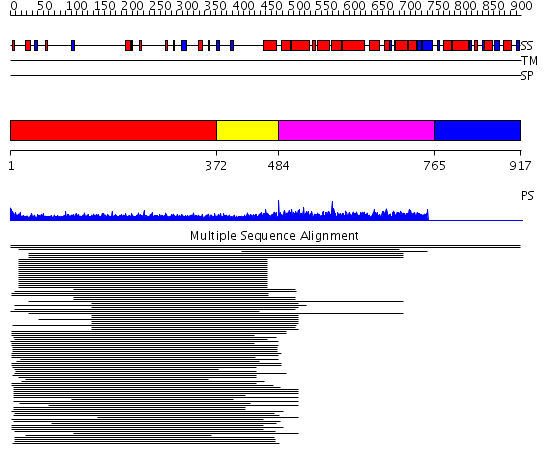
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..371] | 10.56 | RBP1 | |
| 2 | View Details | [372..483] | 11.12 | No description for 1ldvA was found. | |
| 3 | View Details | [484..764] | 5.045757 | Heavy meromyosin subfragment | |
| 4 | View Details | [765..917] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)