













| General Information: |
|
| Name(s) found: |
gi|151945408
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 872 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFDWLNVPG LDLSSGDQAE KRPSNGLGPP SVSFDFGINT AAPHDSSFWD QGSRSHSDTT 60
61 LSYRNNHSNT AADNATNVSS PQKDNPPNGE VRTLSGGDVY AESPEDMQVP LSLSQNQLTH 120
121 EEIRTYLRWY HYICLRTHGK LVRLNDVFRF LTNFNLSQKV KDRIVEIFRS CKNALNIGQF 180
181 FAVLRLVSRA IIYGILPLRR MILEKAPVPK PRPILSSENH EEVYEEVEDD DSSAKTGDQK 240
241 VDFDSFASLL LTGKTTRKRV RRRIKNLNFK SKKVRFSEHI TFQDPPNLNQ ESSNNSEARK 300
301 QDPDAEDEDQ DSNNDSPLDF TLPMDQLLKR LYKRRKNSGL VSSLPSEQQE TEEEKKVLED 360
361 MKDSLSHFKQ IQTVDSASLP ISSVFLQNGN TLPTSNVNNT TVPQQLPLEP LKPTATGSAN 420
421 HLVREEYNQG LHPSNGAIQT GLQPLKPTAT GSANYLMRSH MEQPQSIKPS STPETVTNSG 480
481 GLQPLKPTAT GSANYLMKQH ISPSVNNPVS SMFQAQFTNQ SSSPQSTGPA FLNSPNITLP 540
541 QSNQQQPYQE VNPTQAKIEP SNISPQHTYS NNVRINNGNI VSMPKVEITG AFPPQNTLPQ 600
601 HQQSHLLSPQ NTIPQHQRSQ LISPQNTFTQ NQPILSPQHT YSNNQATMIS PQNTYTNNQQ 660
661 QPQHLPPPPP PRAQQQQQGA IVPPQHMYSN VQKQNNLVPT QPSYTNSPSI QSPNFLSPQN 720
721 AANSYFQSLL SSSPSPNPTP SNASTVNGNN ASNGISSFQN TSAAMNNTQS HQTYIQQQQQ 780
781 QQTQQRIYGG QLSQMQQHPG QLHLNNSDIH SQPNKPNYGM LGQQVHQQQQ QQQQQFPFTA 840
841 DVNRSNSSDI LGNLQSLQQQ VDALQIQYNR RP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
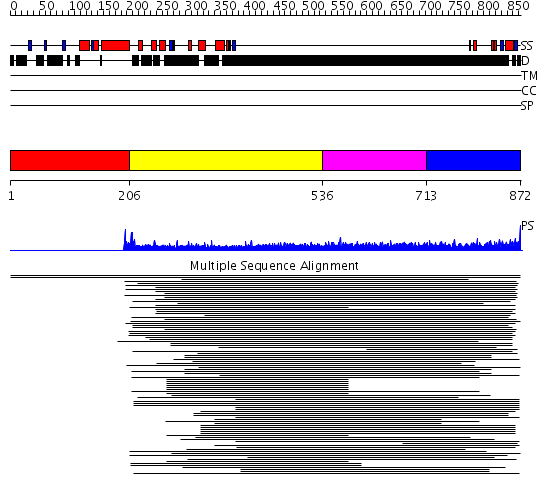
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [206..535] | 4.304997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [536..712] | 2.045757 | Sec24 | |
| 4 | View Details | [713..872] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)