













| General Information: |
|
| Name(s) found: |
ARFS_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1086 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
leaf development
[IGI]
lateral root primordium development [IGI] response to ethylene stimulus [IMP] response to auxin stimulus [IMP][NAS] lateral root development [IGI] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKAPSNGFLP SSNEGEKKPI NSQLWHACAG PLVSLPPVGS LVVYFPQGHS EQVAASMQKQ 60
61 TDFIPNYPNL PSKLICLLHS VTLHADTETD EVYAQMTLQP VNKYDREALL ASDMGLKLNR 120
121 QPTEFFCKTL TASDTSTHGG FSVPRRAAEK IFPPLDFSMQ PPAQEIVAKD LHDTTWTFRH 180
181 IYRGQPKRHL LTTGWSVFVS TKRLFAGDSV LFVRDEKSQL MLGIRRANRQ TPTLSSSVIS 240
241 SDSMHIGILA AAAHANANSS PFTIFFNPRA SPSEFVVPLA KYNKALYAQV SLGMRFRMMF 300
301 ETEDCGVRRY MGTVTGISDL DPVRWKGSQW RNLQVGWDES TAGDRPSRVS IWEIEPVITP 360
361 FYICPPPFFR PKYPRQPGMP DDELDMENAF KRAMPWMGED FGMKDAQSSM FPGLSLVQWM 420
421 SMQQNNPLSG SATPQLPSAL SSFNLPNNFA SNDPSKLLNF QSPNLSSANS QFNKPNTVNH 480
481 ISQQMQAQPA MVKSQQQQQQ QQQQHQHQQQ QLQQQQQLQM SQQQVQQQGI YNNGTIAVAN 540
541 QVSCQSPNQP TGFSQSQLQQ QSMLPTGAKM THQNINSMGN KGLSQMTSFA QEMQFQQQLE 600
601 MHNSSQLLRN QQEQSSLHSL QQNLSQNPQQ LQMQQQSSKP SPSQQLQLQL LQKLQQQQQQ 660
661 QSIPPVSSSL QPQLSALQQT QSHQLQQLLS SQNQQPLAHG NNSFPASTFM QPPQIQVSPQ 720
721 QQGQMSNKNL VAAGRSHSGH TDGEAPSCST SPSANNTGHD NVSPTNFLSR NQQQGQAASV 780
781 SASDSVFERA SNPVQELYTK TESRISQGMM NMKSAGEHFR FKSAVTDQID VSTAGTTYCP 840
841 DVVGPVQQQQ TFPLPSFGFD GDCQSHHPRN NLAFPGNLEA VTSDPLYSQK DFQNLVPNYG 900
901 NTPRDIETEL SSAAISSQSF GIPSIPFKPG CSNEVGGIND SGIMNGGGLW PNQTQRMRTY 960
961 TKVQKRGSVG RSIDVTRYSG YDELRHDLAR MFGIEGQLED PLTSDWKLVY TDHENDILLV1020
1021 GDDPWEEFVN CVQNIKILSS VEVQQMSLDG DLAAIPTTNQ ACSETDSGNA WKVHYEDTSA1080
1081 AASFNR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
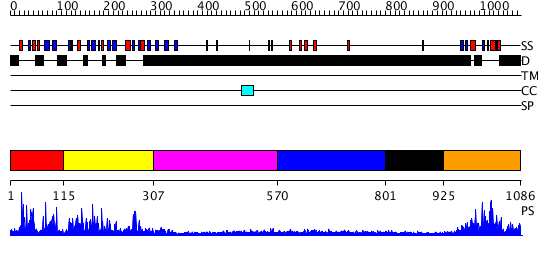
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..306] | 14.39794 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [307..569] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [570..800] | 1.248991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [801..924] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [925..1086] | 2.209981 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)