













| General Information: |
|
| Name(s) found: |
ARFH_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 811 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
flower development
[IGI]
regulation of transcription, DNA-dependent [TAS] response to auxin stimulus [IMP] |
| Molecular Function: |
transcription factor activity
[ISS][TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLSTSGLGQ QGHEGEKCLN SELWHACAGP LVSLPSSGSR VVYFPQGHSE QVAATTNKEV 60
61 DGHIPNYPSL PPQLICQLHN VTMHADVETD EVYAQMTLQP LTPEEQKETF VPIELGIPSK 120
121 QPSNYFCKTL TASDTSTHGG FSVPRRAAEK VFPPLDYTLQ PPAQELIARD LHDVEWKFRH 180
181 IFRGQPKRHL LTTGWSVFVS AKRLVAGDSV IFIRNEKNQL FLGIRHATRP QTIVPSSVLS 240
241 SDSMHIGLLA AAAHASATNS CFTVFFHPRA SQSEFVIQLS KYIKAVFHTR ISVGMRFRML 300
301 FETEESSVRR YMGTITGISD LDSVRWPNSH WRSVKVGWDE STAGERQPRV SLWEIEPLTT 360
361 FPMYPSLFPL RLKRPWHAGT SSLPDGRGDL GSGLTWLRGG GGEQQGLLPL NYPSVGLFPW 420
421 MQQRLDLSQM GTDNNQQYQA MLAAGLQNIG GGDPLRQQFV QLQEPHHQYL QQSASHNSDL 480
481 MLQQQQQQQA SRHLMHAQTQ IMSENLPQQN MRQEVSNQPA GQQQQLQQPD QNAYLNAFKM 540
541 QNGHLQQWQQ QSEMPSPSFM KSDFTDSSNK FATTASPASG DGNLLNFSIT GQSVLPEQLT 600
601 TEGWSPKASN TFSEPLSLPQ AYPGKSLALE PGNPQNPSLF GVDPDSGLFL PSTVPRFASS 660
661 SGDAEASPMS LTDSGFQNSL YSCMQDTTHE LLHGAGQINS SNQTKNFVKV YKSGSVGRSL 720
721 DISRFSSYHE LREELGKMFA IEGLLEDPLR SGWQLVFVDK ENDILLLGDD PWESFVNNVW 780
781 YIKILSPEDV HQMGDHGEGS GGLFPQNPTH L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
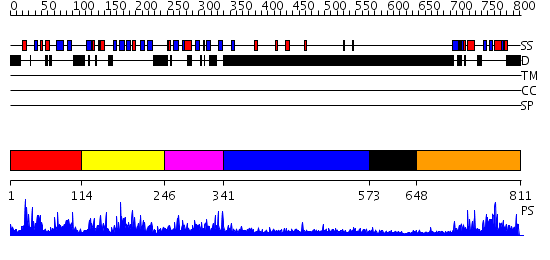
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..113] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [114..245] | 41.221849 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [246..340] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [341..572] | 6.305996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [573..647] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [648..811] | 1.02 | Solution structure of RSGI RUH-024, a PB1 domain in human cDNA, KIAA0049 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)