













| General Information: |
|
| Name(s) found: |
HFM1 /
YGL251C
[SGD]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1046 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
synapsis
[IGI DNA unwinding involved in replication [IDA meiosis [IDA |
| Molecular Function: |
DNA helicase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSEAFPSIY ESNENCIISS PTGSGKTVLF ELAILRLIKE TNSDTNNTKI IYIAPTKSLC 60
61 YEMYKNWFPS FVNLSVGMLT SDTSFLETEK AKKCNIIITT PEKWDLLTRR WSDYSRLFEL 120
121 VKLVLVDEIH TIKEKRGASL EVILTRMNTM CQNIRFVALS ATVPNIEDLA LWLKTNNELP 180
181 ANILSFDESY RQVQLTKFVY GYSFNCKNDF QKDAIYNSKL IEIIEKHADN RPVLIFCPTR 240
241 ASTISTAKFL LNNHIFSKSK KRCNHNPSDK ILNECMQQGI AFHHAGISLE DRTAVEKEFL 300
301 AGSINILCST STLAVGVNLP AYLVIIKGTK SWNSSEIQEY SDLDVLQMIG RAGRPQFETH 360
361 GCAVIMTDSK MKQTYENLIH GTDVLESSLH LNLIEHLAAE TSLETVYSIE TAVNWLRNTF 420
421 FYVRFGKNPA AYQEVNRYVS FHSVEDSQIN QFCQYLLDTL VKVKIIDISN GEYKSTAYGN 480
481 AMTRHYISFE SMKQFINAKK FLSLQGILNL LATSEEFSVM RVRHNEKKLF KEINLSPLLK 540
541 YPFLTEKKQS QIIDRVSQKV SLLIQYELGG LEFPSYEGAS KLHQTLVQDK FLVFRHCFRL 600
601 LKCMVDTFIE KSDGTSLKNT LFLLRSLNGH CWENTPMVLR QLKTIGLVSV RRLIRHGITN 660
661 LEEMGHLSDT QIEYYLNLKI GNGIKIKNDI SLLPCLNIRT KLENCKIENE ELWLTFKVEI 720
721 SATFKSSIWH GQHLSLDIET EKSSGELIDF RRLQVNKLQS PRGFRISAKI SPKLEKIEFS 780
781 IHCQEIAGLG KTIVYSTDHL ASQFSAKTPN IRKDLNSLEK CLFYESSSDG EVGKTSRVSH 840
841 KDGLEESLSS DDSILDYLNE RKKSSKAVES AAVIHPEAHS SSHFSNGRQV RSNGNYECFH 900
901 SCKDKTQCRH LCCKEGIPVK YIKEKGPSSI KPVSKPDQIR QPLLAKNINT TPHLEKRLNS 960
961 KPKQWQEENT DIATVHTLPS KIYNLSQQMS SMEAGEQVLK SGPENCPEII PIDLESSDSY1020
1021 SSNTAASSIS DPNGDLDFLG SDIEFE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
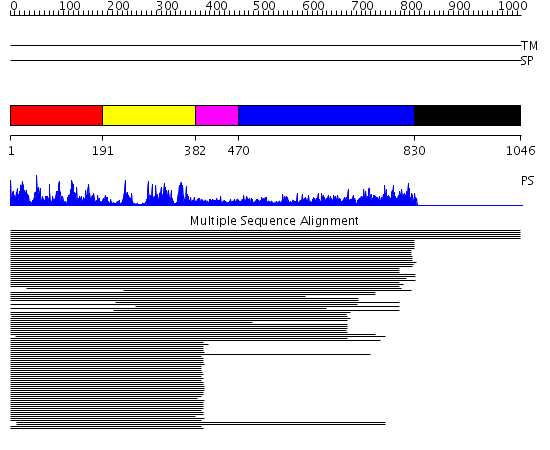
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 79.0 | Putative DEAD box RNA helicase | |
| 2 | View Details | [191..381] | 79.0 | Putative DEAD box RNA helicase | |
| 3 | View Details | [382..469] | 10.154902 | Nucleotide excision repair enzyme UvrB | |
| 4 | View Details | [470..829] | 97.091515 | Sec63 domain No confident structure predictions are available. | |
| 5 | View Details | [830..1046] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)