













| General Information: |
|
| Name(s) found: |
PAPS1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
RNA polyadenylation
[IDA]
|
| Molecular Function: |
nucleotidyltransferase activity
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVQQNGQR FGVSEPISMG GPTEFDVIKT RELEKHLQDV GLYESKEEAV RREEVLGILD 60
61 QIVKTWIKTI SRAKGLNDQL LHEANAKIFT FGSYRLGVHG PGADIDTLCV GPRHATREGD 120
121 FFGELQRMLS EMPEVTELHP VPDAHVPLMG FKLNGVSIDL LYAQLPLWVI PEDLDLSQDS 180
181 ILQNADEQTV RSLNGCRVTD QILRLVPNIQ NFRTTLRCMR FWAKRRGVYS NVSGFLGGIN 240
241 WALLVARICQ LYPNALPNIL VSRFFRVFYQ WNWPNAIFLC SPDEGSLGLQ VWDPRINPKD 300
301 RLHIMPIITP AYPCMNSSYN VSESTLRIMK GEFQRGNEIC EAMESNKADW DTLFEPFAFF 360
361 EAYKNYLQID ISAANVDDLR KWKGWVESRL RQLTLKIERH FKMLHCHPHP HDFQDTSRPL 420
421 HCSYFMGLQR KQGVPAAEGE QFDIRRTVEE FKHTVNAYTL WIPGMEISVG HIKRRSLPNF 480
481 VFPGGVRPSH TSKGTWDSNR RSEHRNSSTS SAPAATTTTT EMSSESKAGS NSPVDGKKRK 540
541 WGDSETLTDQ PRNSKHIAVS VPVENCEGGS PNPSVGSICS SPMKDYCTNG KSEPISKDPP 600
601 ENVVAFSKDP PESLPIEKIA TPQAHETEEL EESFDFGNQV IEQISHKVAV LSATATIPPF 660
661 EATSNGSPFP YEAVEELEVL PTRQPDAAHR PSVQQRKPII KLSFTSLGKT NGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
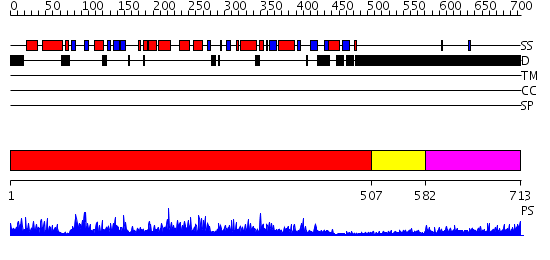
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..506] | 1000.0 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 2 | View Details | [507..581] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [582..713] | 6.212996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)