













| General Information: |
|
| Name(s) found: |
ENO1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
trichome morphogenesis
[IMP]
|
| Molecular Function: |
phosphopyruvate hydratase activity
[IDA][ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALTTKPHHL QRSFLSPSRV SGERYLESAP SCLRFRRSGV QCSVVAKECR VKGVKARQII 60
61 DSRGNPTVEV DLITDDLYRS AVPSGASTGI YEALELRDGD KSVYGGKGVL QAIKNINELV 120
121 APKLIGVDVR NQADVDALML ELDGTPNKSK LGANAILGVS LSVCRAGAGA KGVPLYKHIQ 180
181 ETSGTKELVM PVPAFNVING GSHAGNSLAM QEFMILPVGA TSFSEAFQMG SEVYHTLKGI 240
241 IKTKYGQDAC NVGDEGGFAP NVQDNREGLV LLIDAIEKAG YTGKIKIGMD VAASEFFMKD 300
301 GRYDLNFKKQ PNDGAHVLSA ESLADLYREF IKDFPIVSIE DPFDQDDWSS WASLQSSVDI 360
361 QLVGDDLLVT NPKRIAEAIK KQSCNALLLK VNQIGTVTES IQAALDSKAA GWGVMVSHRS 420
421 GETEDNFIAD LSVGLASGQI KTGAPCRSER LSKYNQLLRI EEELGNVRYA GEAFRSP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
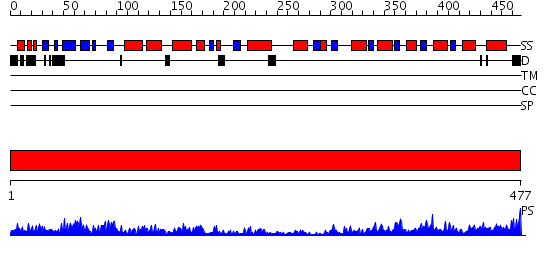
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..477] | 147.0 | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)