













| General Information: |
|
| Name(s) found: |
ARFJ_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
root cap development
[IGI][IMP]
fruit development [IMP] response to carbohydrate stimulus [IMP] regulation of transcription, DNA-dependent [ISS] pattern specification process [IDA] developmental growth [IMP] auxin mediated signaling pathway [IMP] cell division [IGI] petal development [IMP] abscisic acid mediated signaling pathway [IMP] leaf development [IMP] sepal development [IMP] regulation of anthocyanin biosynthetic process [IMP] |
| Molecular Function: |
miRNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQEKSLDPQ LWHACAGSMV QIPSLNSTVF YFAQGHTEHA HAPPDFHAPR VPPLILCRVV 60
61 SVKFLADAET DEVFAKITLL PLPGNDLDLE NDAVLGLTPP SSDGNGNGKE KPASFAKTLT 120
121 QSDANNGGGF SVPRYCAETI FPRLDYSAEP PVQTVIAKDI HGETWKFRHI YRGTPRRHLL 180
181 TTGWSTFVNQ KKLIAGDSIV FLRSESGDLC VGIRRAKRGG LGSNAGSDNP YPGFSGFLRD 240
241 DESTTTTSKL MMMKRNGNND GNAAATGRVR VEAVAEAVAR AACGQAFEVV YYPRASTPEF 300
301 CVKAADVRSA MRIRWCSGMR FKMAFETEDS SRISWFMGTV SAVQVADPIR WPNSPWRLLQ 360
361 VAWDEPDLLQ NVKRVSPWLV ELVSNMPTIH LSPFSPRKKI RIPQPFEFPF HGTKFPIFSP 420
421 GFANNGGGES MCYLSNDNNN APAGIQGARQ AQQLFGSPSP SLLSDLNLSS YTGNNKLHSP 480
481 AMFLSSFNPR HHHYQARDSE NSNNISCSLT MGNPAMVQDK KKSVGSVKTH QFVLFGQPIL 540
541 TEQQVMNRKR FLEEEAEAEE EKGLVARGLT WNYSLQGLET GHCKVFMESE DVGRTLDLSV 600
601 IGSYQELYRK LAEMFHIEER SDLLTHVVYR DANGVIKRIG DEPFSDFMKA TKRLTIKMDI 660
661 GGDNVRKTWI TGIRTGENGI DASTKTGPLS IFA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
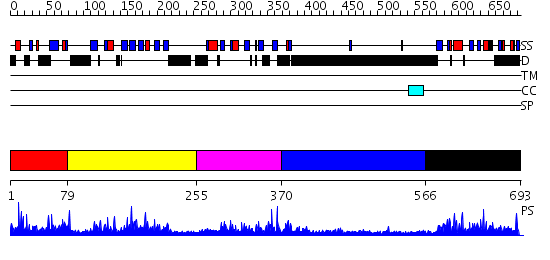
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..78] | 2.078993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [79..254] | 37.045757 | Solution Structure of the B3 DNA-Binding Domain of RAV1 | |
| 3 | View Details | [255..369] | 3.10199 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [370..565] | 3.124996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [566..693] | 1.14 | Solution structure of RSGI RUH-024, a PB1 domain in human cDNA, KIAA0049 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)