













| General Information: |
|
| Name(s) found: |
DRB1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 419 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
[NOT]Cajal body [IDA] nuclear dicing body [IDA] |
| Biological Process: |
response to abscisic acid stimulus
[IMP]
production of miRNAs involved in gene silencing by miRNA [IMP] mRNA cleavage involved in gene silencing by miRNA [IMP] response to cytokinin stimulus [IMP] response to auxin stimulus [IMP] production of ta-siRNAs involved in RNA interference [IMP] |
| Molecular Function: |
miRNA binding
[IDA]
protein binding [IPI] double-stranded RNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSTDVSSGV SNCYVFKSRL QEYAQKYKLP TPVYEIVKEG PSHKSLFQST VILDGVRYNS 60
61 LPGFFNRKAA EQSAAEVALR ELAKSSELSQ CVSQPVHETG LCKNLLQEYA QKMNYAIPLY 120
121 QCQKVETLGR VTQFTCTVEI GGIKYTGAAT RTKKDAEISA GRTALLAIQS DTKNNLANYN 180
181 TQLTVLPCEK KTIQAAIPLK ETVKTLKARK AQFKKKAQKG KRTVAKNPED IIIPPQPTDH 240
241 CQNDQSEKIE TTPNLEPSSC MNGLKEAAFG SVETEKIETT PNLEPPSCMN GLKEAAFGSV 300
301 ETEKIETTPN LEPPSCMNGL KEAAFGSVET EKIETTPNLE PSSCMNGLKE AAFGSVETEK 360
361 IETTPNLEPP SCMNGLKEAA FGSVETEKIE TTPNLESSSC MSGLKEAAFG SVETEASHA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
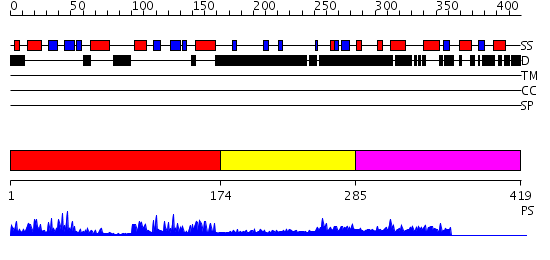
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 22.0 | dsRNA-dependent protein kinase pkr | |
| 2 | View Details | [174..284] | 1.172989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [285..419] | 4.0 | Crystal structure of the catalytic domain of an adenosine deaminase that acts on RNA (hADAR2) bound to inositol hexakisphosphate (IHP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)