













| General Information: |
|
| Name(s) found: |
gi|25373341
[NCBI NR]
gi|15223895 [NCBI NR] gi|11079482 [NCBI NR] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTTQLFKGA NVFMSRNLVP PEVFDTLLDA FKLNGAEIFL CCDPSRSGPS DFHVIASPDH 60
61 EKFKDLKAKG CNLIGPQCAL FCAKEGRPLP QRGFTCCLAM DGLKVLASGF LVDEKVKIKE 120
121 LVTSMGGVLL SRASSDVNFV IVKNVLAAKY KWALNKKPIV TLNWLHRCWN EHRVVPQEPY 180
181 KIPPFSGLTI CVTRIPAGDK YKVARKWGHI QIVTRKWFQQ SIDKKVCLNE ESYPVLGSIP 240
241 LTRGVRDLGV HNGLEKFPSA ATASAADSYV SCAQSRDSDI EASASQNVFP TSMNPSTDVK 300
301 EPGGGPTARP QEQNIDGCTA RDSESEDNDL YLSDCRIFLL GFEASEMRKL AKLVRRGGGS 360
361 RYMLLNERMT HIVVGTPSER EARSVAASGV IQVVIPSWLE DCDREKKEIP VHNIYTANHL 420
421 ILPRDSACLT KGSFARMSSM EQTKNTHDQT MVGCLLAVSS HILYSPLPCQ TPLPGFESLC 480
481 ICSSQHNEKN VELLRNLSVV LGADFVERLT RKVTHLICNF AKGDKYVRAS KWGIISVTPD 540
541 WLYECVRQNQ VVCTDNFHPR ELTTQDREAG SQFHTQFVPM ASRDSMSLPV SHSEDREKIQ 600
601 SFAGKSGCGK GEVYNRLGEI GKEQTFPSKK AKLLRDGQES DVFPVRELPS NCDRPSHSGD 660
661 GIVTGYDVAS GREVPDVADT IEDLLEQTSK IQDQKSPGRI LEKTVSLNEQ YNTGNHSVTG 720
721 LSRHWINRVH KNDDMGSPPG DATTDTYGNF SETQTESQVV GYEEDLSGRQ MLIDRVRTRS 780
781 SLT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
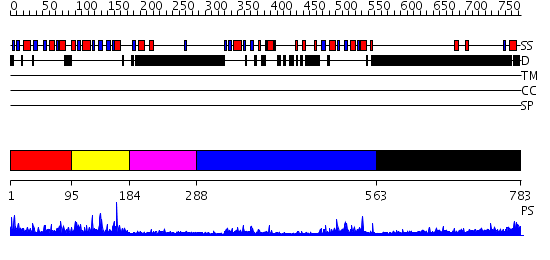
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | 1.87 | Co-Crystal Structure of Lif1p-Lig4p | |
| 2 | View Details | [95..183] | 19.69897 | No description for 2d8mA was found. | |
| 3 | View Details | [184..287] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [288..562] | 22.522879 | No description for 2owoA was found. | |
| 5 | View Details | [563..783] | 5.172996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)