













| General Information: |
|
| Name(s) found: |
CPL2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 771 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
response to osmotic stress
[IMP]
developmental growth [IMP] response to auxin stimulus [IMP] |
| Molecular Function: |
double-stranded RNA binding
[ISS]
phosphatase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRLGHKSVV YHGDLRLGEL DVNHVSSSHE FRFPNDEIRI HHLSPAGERC PPLAILQTIA 60
61 SFAVRCKLES SAPVKSQELM HLHAVCFHEL KTAVVMLGDE EIHLVAMPSK EKKFPCFWCF 120
121 SVPSGLYDSC LRMLNTRCLS IVFDLDETLI VANTMKSFED RIEALKSWIS REMDPVRING 180
181 MSAELKRYMD DRMLLKQYID NDYAFDNGVL LKAQPEEVRP TSDGQEKVCR PVIRLPEKNT 240
241 VLTRIKPEIR DTSVLVKLRP AWEELRSYLT AKTRKRFEVY VCTMAERDYA LEMWRLLDPE 300
301 AHLISLKELR DRIVCVKPDA KKSLLSVFNG GICHPKMAMV IDDRMKVWED KDQPRVHVVS 360
361 AYLPYYAPQA EACALVVPHL CVARNVACNV RGYFFKEFDE SLMSSISLVY YEDDVENLPP 420
421 SPDVSNYVVI EDPGFASNGN INAPPINEGM CGGEVERRLN QAAAADHSTL PATSNAEQKP 480
481 ETPKPQIAVI PNNASTATAA ALLPSHKPSL LGAPRRDGFT FSDGGRPLMM RPGVDIRNQN 540
541 FNQPPILAKI PMQPPSSSMH SPGGWLVDDE NRPSFPGRPS GLYPSQFPHG TPGSAPVGPF 600
601 AHPSHLRSEE VAMDDDLKRQ NPSRQTTEGG ISQNHLVSNG REHHTDGGKS NGGQSHLFVS 660
661 ALQEIGRRCG SKVEFRTVIS TNKELQFSVE VLFTGEKIGI GMAKTKKDAH QQAAENALRS 720
721 LAEKYVAHVA PLARETEKGP ENDNGFLWES SEDVSNKGLE EEAPKENISE L |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
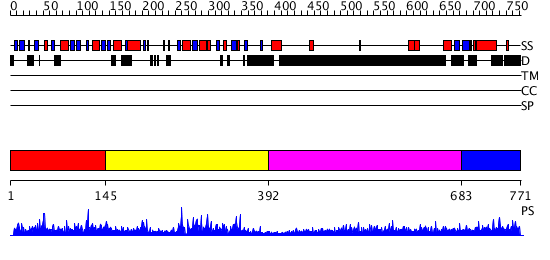
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [145..391] | 43.39794 | No description for 2ghqA was found. | |
| 3 | View Details | [392..682] | 3.69897 | No description for 1y0fA was found. | |
| 4 | View Details | [683..771] | 1.54 | Crystal structure of rnt1p dsRBD |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)